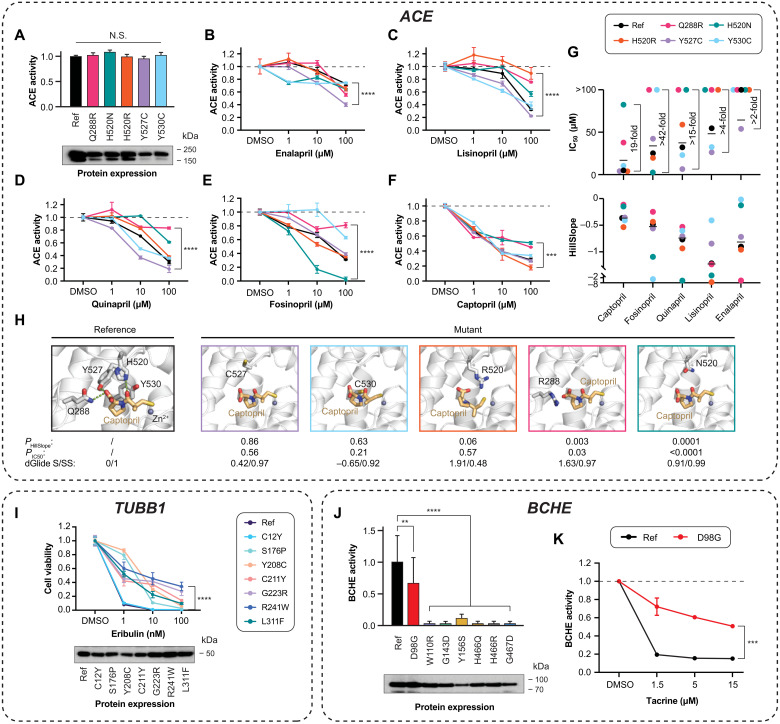
Fig. 4. Naturally occurring drug-binding site variability affects drug response in vitro.
Effects of binding site variants in ACE (A to H), TUBB1 (I), and BCHE (J and K) were evaluated using functional assays (see Materials and Methods). (A) None of the ACE drug-binding site variants had major impacts on ACE expression or baseline ACE activity. Data are shown as means ± SEM; n = 3. Dose-response curves of reference ACE and its variants are shown to the clinically approved ACEis enalapril (B), lisinopril (C), quinapril (D), fosinopril (E), and captopril (F). ***P < 0.001; ****P < 0 (F test). (G) The IC50 values and HillSlope coefficients are shown for each ACE variant–ACEi pair, and the largest fold change across variants is indicated for each drug. (H) Cocrystallized structure of captopril binding to ACE [Protein Data Bank (PDB) ID: 1UZF] and docking poses of captopril with the five ACE variant structures are shown. Atom color code in sticks: oxygen (red), nitrogen (blue), and sulfur (yellow). The zinc ion and hydrogen bonds are shown as violet sphere and green dashed lines, respectively. Parameters indicating the differences between wild-type and variants are shown below the structures. (I) Eribulin effect in cells transfected with reference TUBB1 or naturally occurring TUBB1 variants. Note that viability in variant carriers is strongly increased indicating eribulin resistance. (J) With the exception of D98G, drug-binding site variants in BCHE-abrogated enzymatic function (**P < 0.01; ****P < 0.0001; heteroscedastic two-tailed t test; n = 3). (K) Inhibitory effects of the cholinesterase inhibitor tacrine were strongly reduced in D98G compared to reference enzyme (***P < 0.001; F test). N.S., not significant; DMSO, dimethyl sulfoxide.