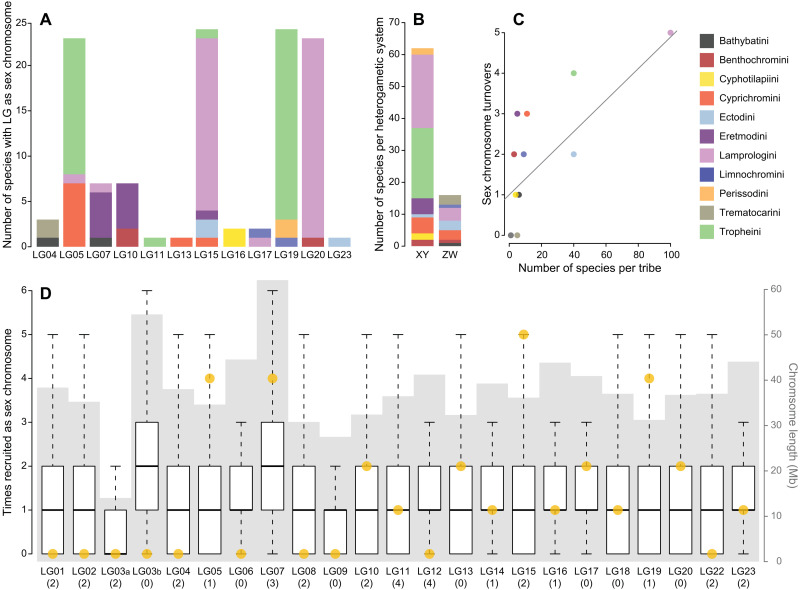
Fig. 2. Nonrandom sex chromosome distribution in LT cichlids.
(A) Use of different LGs as sex chromosomes. Bars represent the number of times a reference genome LG has been detected as sex-linked at the species level with colors referring to tribe (permissive dataset). (B) The occurrence of SD systems. Bars represent how often an XY or ZW heterogametic SD system was identified at the species level (permissive dataset) and with colors referring to tribe. (C) Association between species richness and sex chromosome turnover. The number of sex chromosome turnovers leading to the tips of each tribe (permissive dataset) is associated with the number of species investigated in each tribe (pGLS, P = 0.0043, coefficient = 0.039). Dots are colored according to tribes; the line represents the linear model fitted to the data. (D) Boxplots showing the expected number of sex chromosome recruitments if recruitment was at random (10,000 permutations). Boxplot centerlines represent the median, box limits the upper and lower quartiles, and whiskers the 1.5× interquartile range. Outliers are not shown. Ten reference LGs were never implicated in a turnover event in LT cichlids. Under random recruitment in the simulations, this pattern occurred only in 2.01% of all simulations. Yellow dots indicate the number of observed sex chromosome recruitments per reference genome LG derived from ancestral state reconstructions (permissive dataset), gray background shading represents chromosome length in megabases derived from the reference genome, and numbers below each boxplot indicate the number of previously described sex-determining genes on these reference genome LGs.