FIGURE 2.
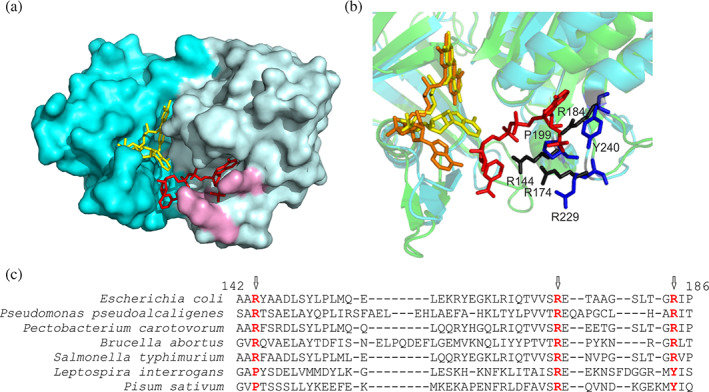
The nucleotide binding site of FNR. (a) Surface model of EcFPR (PDB 2XNJ) containing bound NADP+. FAD is represented in yellow and NADP+ in red. The three arginine residues that interact with NADP+ are shown in purple. (b) Crystal structure of EcFPR (light blue) superimposed to PeaFNR (PDB 1QG0) (green). Arginines present in EcFPR are shown in black and amino acids corresponding to PeaFNR in blue. The NADP+ molecule of EcFPR is shown in red. The FAD molecules have different conformation: folded (yellow) in EcFPR and extended (orange) in PeaFNR. (c) Structural alignment of different FPRs and of the plastidic type LepFNR and PeaFNR. The three arginine residues would be conserved in the FPRs, but only one would be present in the plastidic type (colored in red). Panels A and B were designed from the structures of the proteins obtained by X‐ray diffraction, using the Pymol version 1.8.0.0. program. Panel C alignment was performed with the T‐Coffee Software. The results are consistent with the Pymol alignment function
