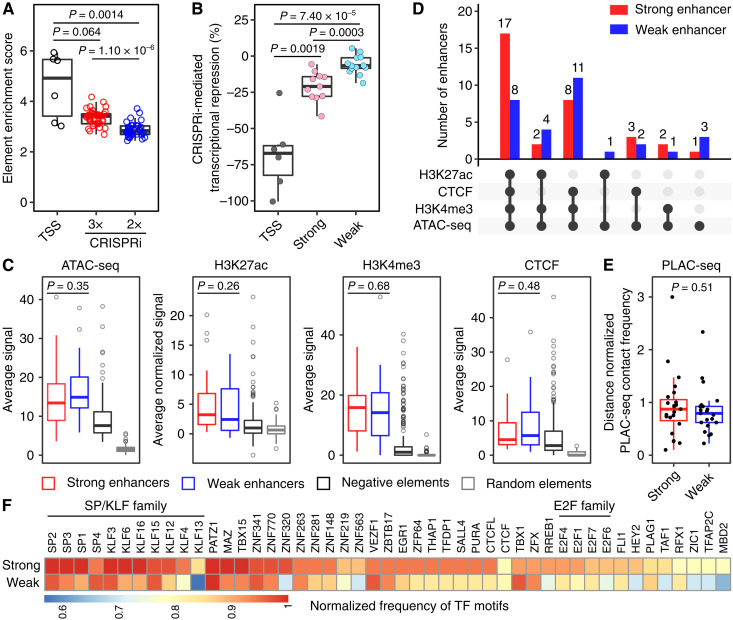
Fig. 4. CRISPRpath can distinguish weak and strong enhancers by imposing different selection pressures.
(A) Box plots show the enrichment score of the tested elements. TSS regions (black circles) show highest enrichment scores. Enhancers uniquely identified from the lower selection pressure (CRISPRi 2×, blue circles) exhibit lower enrichment scores compared to the enhancers identified from the higher selection pressure (CRISPRi 3×, red circles). Each circle represents an individual element. P values are from Wilcoxon test. (B) Box plots show that the CRISPRi perturbation at enhancers induced various degrees of transcriptional repression of target genes measured with RT-qPCR. Each dot represents the average value from three biological replicates. CRISPRi targeting TSS regions (dark gray) achieved the highest transcriptional repression. CRISPRi targeting strong enhancers (pink) leads to a more substantial transcription silencing of the target gene compared to CRISPRi targeting weak enhancers (cyan). P values are from Wilcoxon test. (C) Enrichment analysis of ATAC-seq, H3K27ac, H3K4me3, and CTCF binding signals for strong (n = 33) and weak (n = 30) enhancers. P values for the difference between strong and weak enhancers are from Wilcoxon test; see table S7 for P values of all pairwise comparisons. Box plots (A to C) indicate the median, IQR, Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. (D) Intersection of genomic features for weak enhancers (blue bar) and strong enhancers (red bar). (E) Distance normalized H3K4me3 PLAC-seq contact frequency for strong (n = 23) and weak (n = 21) enhancers. Only the enhancers for HPRT1, MLH1, PMS2, and PCNA are included (see Materials and Methods for details). Box plots indicate the median, IQR, Q1 − 1.5 × IQR, and Q3 + 1.5 × IQR. P value is from Wilcoxon test. (F) Heatmap shows the normalized frequency of transcription factor (TF) motifs found in strong and weak enhancers.