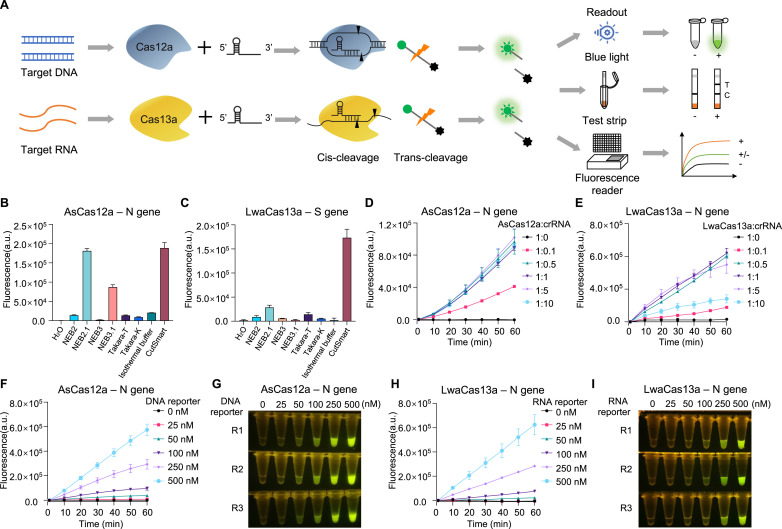
Fig. 1.
Optimization of key parameters for CRISPR detection systems.
(A) Schematic description of CRISPR-Cas12a/Cas13a detection systems for nucleic acids.
(B, C) Comparison of the indicated commercial reaction buffers for their effects on AsCas12a- and LwaCas13a-mediated trans-cleavage activity. A synthetic DNA template corresponding to SARS-CoV-2 N gene (B), and a synthetic RNA template corresponding to SARS-CoV-2 S gene (C) are used. (D, E) Evaluation of the indicated molecular ratio of Cas protein:crRNA for the effect on AsCas12a detection (D) and LwaCas13a detection (E). (F–
I) The effect of indicated amount of fluorescence reporter on CRISPR detection assays using either AsCas12a (F and G) or LwaCas13a (H and I). The results are shown by either real-time recording of fluorescence signal (F and H) or endpoint (60
min) visualization (G and I) under blue light illuminator. R1, R2 and R3 indicate three replicates. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)