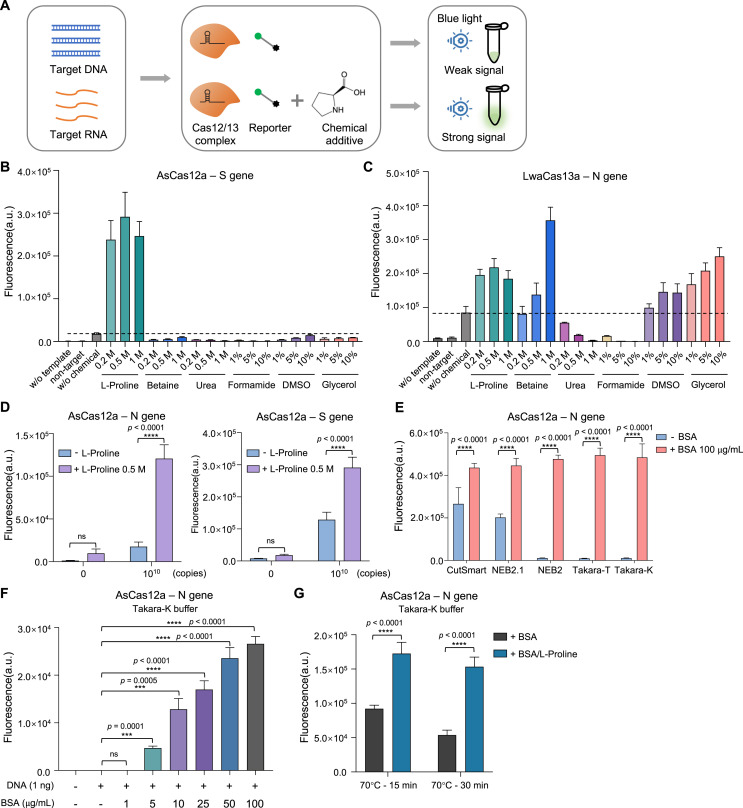
Fig. 3.
Effects of chemical additives on CRISPR detection phase
(A) Schematic diagram showing the workflow to evaluate chemical additive effect on CRISPR detection system. (B, C) Endpoint (60
min) recording of fluorescence detection signals for either AsCas12a (B) or LwaCas13a (C) system. w
/o template means no input; non-target means non-targeting template (N gene template as S gene non-target, and vice versa); w/o chemical means no chemical addition. (D) Comparison of fluorescent signals resulted from AsCas12a-based detection with or without L-proline addition. Two-way ANOVA test, ****p < 0.0001; ns means not significant. (E) The effect of BSA addition on AsCas12a-based detection system. Two-way ANOVA test, ****p < 0.0001. (F) Comparison of different concentrations of BSA addition for the effects on AsCas12a-mediated detection. Unpaired two-tailed t-test, ***p < 0.001, ****p < 0.0001. (G) Evaluation of L-proline's effect on protecting BSA from heat-induced denature and BSA's capability to enhance AsCas12a detection. BSA along or BSA co-incubated with L-proline are treated at 70 °C for 15 min or 30 min, before serving as additive in AsCas12a assays. Two-way ANOVA test, ****p < 0.0001. Error bars represent mean ± s.d. (n = 3). a. u., arbitrary unit.