Figure 6.
TRIM8 interacts with and ubiquitinates EWS/FLI in Ewing sarcoma cells
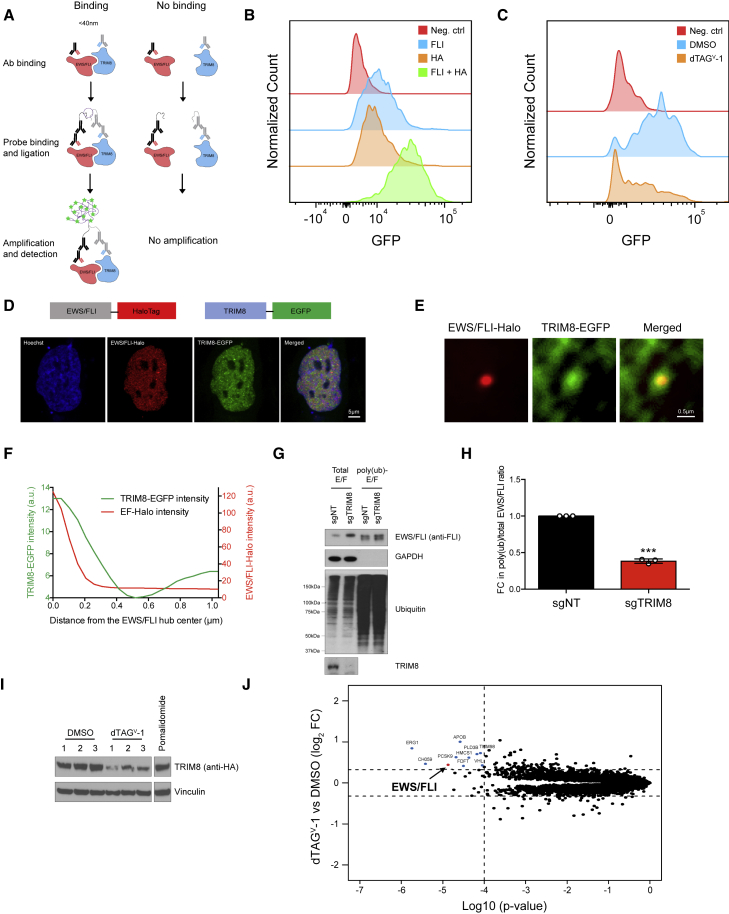
(A) Schematic of the proximity ligation assay (PLA).
(B) Histogram showing fluorescence signal from the PLA in TC71 C-dTAG TRIM8 cells. No antibody (neg. ctrl), anti-FLI, and anti-HA alone were used as negative controls. Anti-FLI and anti-HA combination were used to detect interaction between TRIM8 and EWS/FLI.
(C) PLA signal TC71 C-dTAG TRIM8 cells treated with either DMSO or dTAGV-1 (1 μM) for 48 h. Data representative of three independent experiments.
(D) Schematic of fluorescent proteins expressed in A673-E/F-Halo cell line is shown above. Airyscan super-resolution images of the nucleus (Hoechst stained, blue), endogenous EWS/FLI-Halo (JF646 labeled, red), and transiently expressed TRIM8-EGFP (green) in an A673-E/F-Halo cell.
(E) Two-color image showing that endogenous EWS/FLI-Halo hubs enrich for TRIM8-EGFP. The nuclear fluorescence background is subtracted in both channels of the average image (see the STAR Methods for details).
(F) Average background-free radial profiles of EWS/FLI-Halo and TRIM8-EGFP at EWS/FLI-Halo hubs (averaged from 6,872 hubs in 15 cells).
(G and H) Poly-ubiquitinated proteins in TC71 cells infected with either non-targeting or TRIM8-targeting sgRNA were enriched with tandem ubiquitin binding entities (TUBE). Both input and TUBE-enriched proteins were immunoblotted with the indicated antibodies. EWS/FLI levels were quantified using ImageJ and the poly(ub)-E/F:total-E/F ratio was used to calculate poly-ubiquitinated EWS/FLI levels. Fold change (FC) in poly-ubiquitinated EWS/FLI ±SEM from three independent experiments is shown in (H). ∗∗∗p < 0.001.
(I) TC71 C-dTAG TRIM8 cells treated with 1 μM of dTAGV-1 for 6 h in triplicate were lysed and immunoblotted with the indicated antibodies. Six hour pomalidomide (1 μM) treatment served as a positive control for TMT mass spectrometry.
(J) Total proteome of cells in (I) was analyzed using TMT quantification mass spectrometry. Non-EWS/FLI proteins that significantly changed in abundance are highlighted in blue and EWS/FLI in red. Significance cutoffs used were p < 0.0001 and fold change (FC) > 1.25 and noted with dotted lines. Note: ERG1 (Q14534) is a squalene monooxygenase and not the ETS family member ERG (P11308).