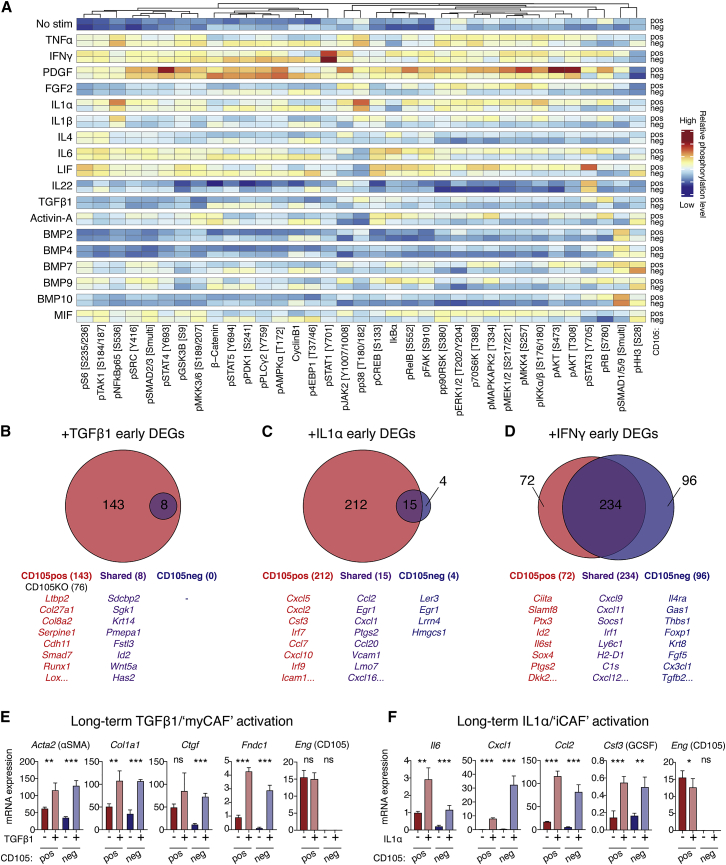
Figure 5.
Differential signaling engagement of CD105pos and CD105neg PaFs
(A) MC analysis of CD105pos and CD105neg PaFs signaling. Data are displayed as median mass intensities (MMI) and column Z scores. Specific phosphorylation sites are annotated in brackets.
(B–D) RNA-seq analysis of CD105pos and CD105neg PaFs stimulated as displayed for 6 h (n = 3). DEGs were identified using DEseq2 with Benjamini-Hochberg adjusted p < 0.05. Data are displayed as Venn diagrams (top), with example genes listed (below). Unique DEGs of CD105pos PaFs in red, CD105neg PaFs in blue, and shared in purple. Numbers of significant DEGs are displayed in parenthesis.
(E and F) Expression of myCAF (E) and iCAF (F) genes from CD105pos and CD105neg PaFs stimulated with TGF-β1 or IL-1α (n = 4) for 3 days. Eng expression is also shown. Data displayed as mean ± SD.
Samples were compared using unpaired t tests (E and F). ns, not significant; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.