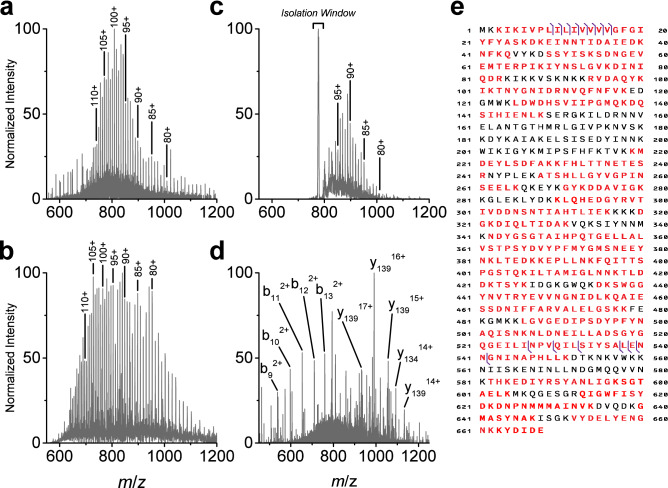
Figure 1.
Identification of PBP2amecA protein from MRSA cell extract by LC–MS/MS. (a) Intact wild-type PBP2amecA MS spectrum from a representative MRSA strain (ATCC 33,591) separated by LC. Intact protein precursor ion charge states are labelled from 80 to 110. Data are representative for multiple MRSA strains (n ≥ 3). (b) Recombinant His6-PBP2amecA MS spectrum acquired using direct infusion. Identical precursor ion charges states are labelled as wild-type PBP2amecA spectrum. (c) Intact wild-type PBP2amecA MS spectrum produced during LC separation of cell extract and PTCR-mediated separation of superposed protein ion populations. Isolation window was centered at m/z 777 with a width of 5 m/z. Precursor ion charge states are labelled from 80 to 95. Data are representative of multiple technical replicates from the same MRSA cell extract (ATCC 33,591). (d) MS/MS spectrum of intact wild-type PBP2amecA precursor ion at m/z 793 (charge state = 102), using 1.5 m/z isolation window and fragmented with an HCD collision energy of 10 eV. Abundant N-terminal (b-ions) and C-terminal (y-ions) fragment ions are labelled. Fragmentation data are representative for multiple charge states and different MRSA strains (n ≥ 100). (e) Associated b- and y-ion fragment location for MS/MS of intact wild-type PBP2amecA protein indicated by purple vertical lines. Red colored amino acids indicate protein coverage (72.2%) for complementary bottom-up peptide analysis and protein characterization. Protein coverage associated with peptide data are from multiple analyses (n = 12).