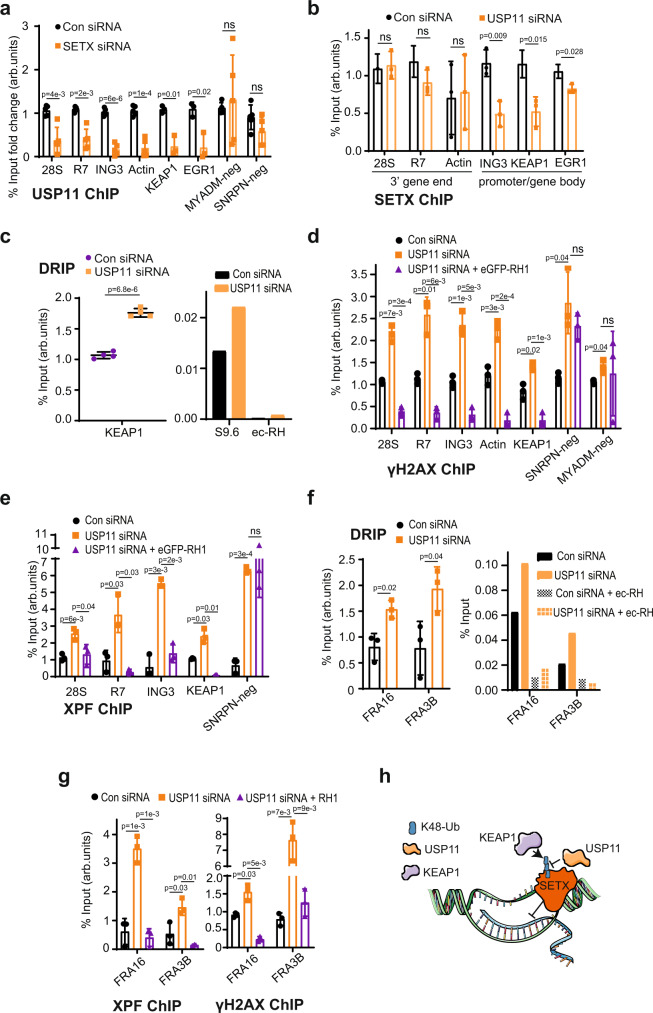
Fig. 7. R-loops formed upon USP11 loss are processed to double-strand breaks.
a Control siRNA (Con) or SETX siRNA-treated HEK-293 cells were subjected to a USP11 ChIP followed by qPCR using primers targeting nucleolar (28S, R7) and nuclear (ING3, Actin, KEAP1, EGR1, SNRPN-neg, MYADM-neg) loci and presented as % input fold change compared to Con. Data represent the average ± SD from five biological repeats. ns; p > 0.05, two-tailed Student’s t-test. b Control siRNA (Con) or USP11 siRNA-treated HEK-293 cells were subjected to a SETX ChIP followed by qPCR using primers targeting nucleolar (28S, R7) and nuclear (ING3, Actin, KEAP1, EGR1) loci. Data represent the average ± SD from three biological repeats. ns; p > 0.05, two-tailed Student’s t-test. c Lysates from Control (Con siRNA) and USP11-depleted cells (USP11 siRNA) were subjected to a DNA/RNA immunoprecipitation (DRIP) protocol using S9.6 antibodies. Quantitative PCR was conducted using primers targeting nuclear KEAP1 locus. Pooled repeats (left panel) and raw % input values from a representative experiment are shown (right panel), and data represent the average ± SD from four biological repeats. p-Values calculated using two-tailed Student’s t-test. d HEK-293 cells were transfected with Control siRNA, USP11 siRNA and eGFP-RNase H1 (eGFP-RH1), and subsequently subjected to a γH2AX ChIP followed by qPCR using primers targeting nucleolar (28S, R7) and nuclear (ING3, Actin, KEAP1, SNRPN-neg, MYADM-neg) loci. Data represent the average ± SD from three biological repeats. ns; p > 0.05, two-tailed Student’s t-test. e HEK-293 cells were transfected with Control (Con) siRNA, USP11 siRNA and eGFP-RNase H1 (eGFP-RH1), and subsequently subjected to a XPF ChIP followed by qPCR using primers targeting nucleolar (28S, R7) and nuclear (ING3, KEAP1, SNRPN-neg) loci. Data represent the average ± SD from three biological repeats. ns; p > 0.05, two-tailed Student’s t-test. f Lysates from Control (Con) siRNA and USP11-depleted cells (USP11 siRNA) were subjected to a DNA/RNA immunoprecipitation (DRIP) protocol using S9.6 antibodies. Quantitative PCR was conducted using primers targeting nuclear common fragile sites FRA16 and FRA3B. In vitro, on-bead ec-RNase-H (ec-RH) treatment served as a signal validation control. Pooled repeats (left panel) and raw % input values from a representative experiment are shown (right panel), and data represent the average ± SD from three biological repeats. p-Values calculated using two-tailed Student’s t-test. g HEK-293 cells were transfected with Control (Con) siRNA, USP11 siRNA and eGFP-RNase H1 (RH1), and subsequently subjected to a XPF ChIP (left panel) and γH2A.X ChIP (right panel) followed by qPCR using primers targeting nuclear common fragile sites FRA16 and FRA3B. Data represent the average ± SD from three biological repeats. p-Values calculated using two-tailed Student’s t-test. h A model depicting R-loop regulation by SETX-USP11-KEAP1 axis. SETX protein level is regulated via ubiquitination by KEAP1 and deubiquitination by USP11. We suggest that the extent of SETX binding to USP11 and KEAP1 is controlled to favour more binding to USP11, thus increasing SETX levels, or more binding to KEAP1, thus reducing SETX level, at distinct genomic loci. The spatial regulation and control of SETX levels would ensure a fine balance to favour physiological R-loops that are required to promote transcription and, at the same time, suppress pathological R-loops that cause genomic instability.