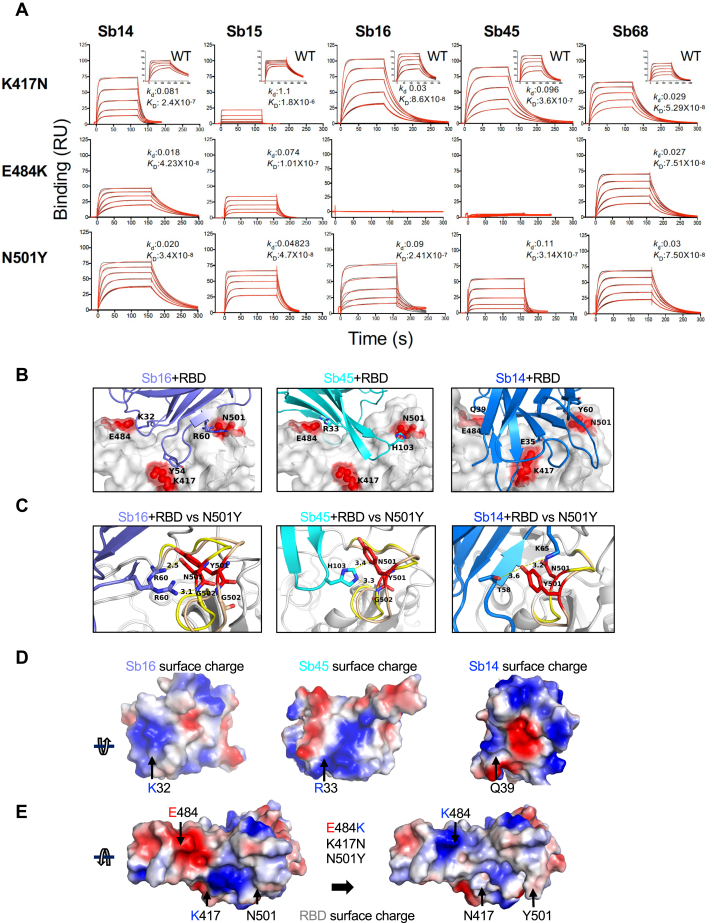
Figure 6.
RBD mutations affect sybody binding.A, SPR binding of each of the indicated sybodies (across top) to each of the individual RBD mutants. Inset shows binding of sybodies to the WT RBD (from Fig. 1). Experimental tracings are shown in red, curve fits in black, and kd (s−1) and KD (M) values as determined from global fitting with BIAeval 2.0 are provided in each panel. B, location of contacts of Sb16, Sb45, and Sb14 is shown. E484, K417, and N501 of the RBD (WT) interact with K32, Y54, and R60 of Sb16, respectively; E484 and N501 of the RBD (WT) interact with R33 and H103 of Sb45, respectively; and E484, K417, and N501 of the RBD (WT) interact with Q39, E35, and Y60 of Sb14, respectively. C, comparison of complex structures with minimized models involving the N501Y mutation. In silico mutagenesis of N501Y was performed using 7KGK (Sb16+RBD), 7KGJ (Sb45+RBD), and 7MFU (Sb14+RBD+Sb68). After amino acid substitution in Coot, local energy minimization (within 15–20 Å of the mutant residue) was performed through three rounds in PHENIX. For the Sb16–RBD complex, when N501 is mutated to Y501, the loop (496–506, from yellow to wheat) extends about 2.4 Å, but R60 (revealing a double conformation) still forms hydrogen bonds with the Y501 loop; for the Sb45–RBD complex, when N501 is mutated to Y501, the loop (496–506, from yellow to wheat) extends about 1.0 Å, but H103 of Sb45 would still interact with Y501; for the Sb14–RBD complex, when N501 is mutated to Y501, the loop (496–506, from yellow to wheat) is extended about 2.0 Å, but T58 and K65 still form hydrogen bonds with Y501. D, the surface charge of Sb16; K32 forms a hydrogen bond with E484 of the RBD with the opposite charge; the surface charge of Sb45, R33 forms a hydrogen bond with E484 of the RBD with the opposite charge; the surface charge of Sb14, Q39 (a neutral residue) interacts with E484 of the RBD. E, surface charge of the WT RBD and surface charge of the RBD with the three mutations (E484, K417N, and N501Y). When E484 is mutated to K484, the surface charge is changed from negative to positive. Therefore, the hydrogen bonds are broken, pushing Sb16 and Sb45 out of contact, whereas because Q39 of Sb14 is not a charged residue, it still may interact with K484 of the mutated RBD. RBD, receptor-binding domain; SPR, surface plasmon resonance.