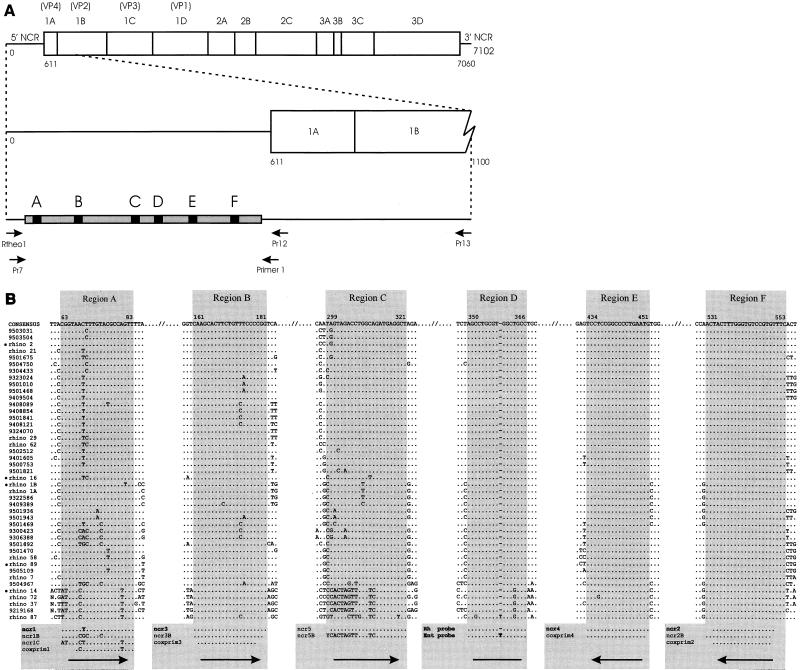
FIG. 1.
(A) Locations and orientations (arrows) of primers used to amplify the 5′ NCRs of 39 rhinovirus isolates and prototypes (see Materials and Methods). The gray bar represents the 5′ NCR from which the nucleotide sequence was determined. The black regions, labeled A to F, indicate the locations of the relatively long, well-conserved subregions. For reference, the complete rhinovirus genome is given at the top. (B) Nucleotide sequences (cDNA) of the six relatively long, well-conserved subregions of the rhinovirus 5′ NCR. At the top, the consensus sequence of each region is given together with its relative position based on the rhinovirus serotype 2 sequence. Identical residues are indicated with dots; a dash indicates that no nucleotide residue is present at that particular position. At the bottom, the sequences of the primers and probes used in the study are given. Arrows indicate primer orientations. The coxprim1 to coxprim4 primers were previously published by Kämmerer et al. (22). The codes of the rhinovirus field isolates and prototypes from which the sequences were derived are given on the left. Rhinovirus serotype x is abbreviated as rhino x. The seven-digit numbers represent field isolates; the first two digits indicate the year of isolation. The sequences of rhinoviruses marked with bullets were downloaded from the GenBank sequence database.