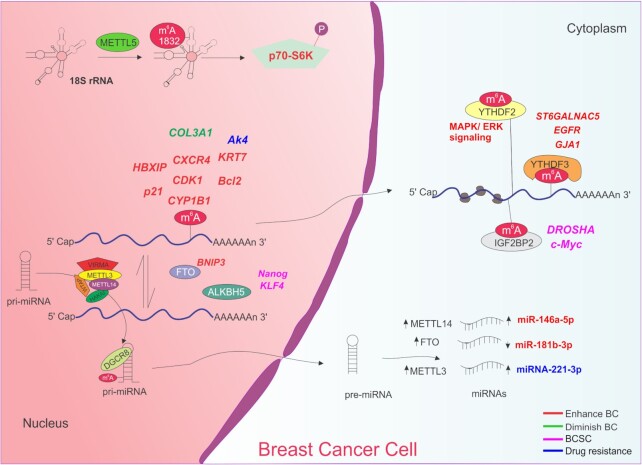
Figure 5.
Functions of m6A in breast cancer. m6A modification is dynamically regulated by its writers (METTL3, METTL14, WTAP, HAKAI and VIRMA) erasers (FTO and ALKBH5) and readers (YTHDFC3 and IGF2BP2). Only enzymes with reported function in breast cancer are depicted for simplicity. Deposition of m6A on distinct transcripts can enhance (HBXIP, p21, CXCR4, CDK1, CYP1B1, KRT7, Bcl2; red) or diminish (COL3A1; green) breast cancer initiation and progression and affect therapy outcome (AK4; blue). Demethylation of BNIP3 also enhances breast cancer (red) whereas demethylation of Nanog, KLF4 and Sox2 promotes the breast cancer stem cell phenotype. Similarly, recognition of m6A-modified DROSHA and c-Myc mRNAs leads to the acquisition of breast cancer stem cell characteristics. YTHDFC3 binds to ST6GALNAC5, EGFR and GJA1 to induce breast cancer metastasis. YTHDF2 recognizes mRNAs involved in MAPK/ERK signaling. Deposition of m6A on pri-miRNA allows DGCR8 recognition and further processing. Increased expression of METTL14, FTO and METTL3 alters the expression of miRNA, enhancing the breast cancer phenotype (red) or induces drug resistance (blue) in breast cancer cells. Enhanced METTL5 mediated methylation at adenosine 1832 of mammalian 18S rRNA promotes p70-S6K activation and an increased translation initiation thus stimulating breast cancer cell growth.