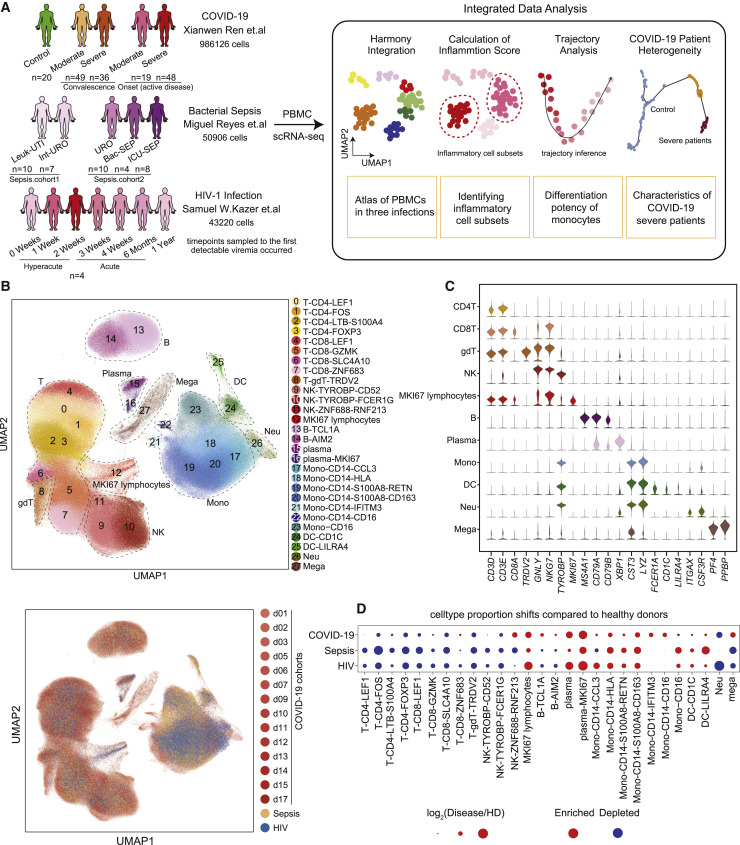
Figure 1.
Single-cell transcriptional landscape of COVID-19, sepsis, and HIV infection
(A) A schematic outline depicting the workflow for data collection from published literature and subsequent integrated analysis. Numbers indicate the number of samples of different cohorts (COVID-19, sepsis, HIV infection) and the number of single-cell transcriptomes analyzed.
(B) Top, uniform manifold approximation and projection (UMAP) embeddings of integrated single-cell transcriptomes of COVID-19, sepsis, HIV infection, and healthy donors’ (HDs’) samples. Cells are colored by cell subtypes, and dashed circles indicate the major cell types. Bottom, UMAP embeddings of different datasets illustrating no obvious batch effect in this integrated atlas.
(C) Violin plots of canonical markers (columns) for major cell types (rows).
(D) Dot plots of the proportion shifts of cell subtypes in COVID-19, sepsis, and HIV, compared to HDs. The circle color indicates enrichment (red) or depletion (blue), and the circle size indicates the values of log2(Disease/HD). Mono, monocytes; DCs, dendritic cells; Neu, neutrophils; Mega, megakaryocytes.