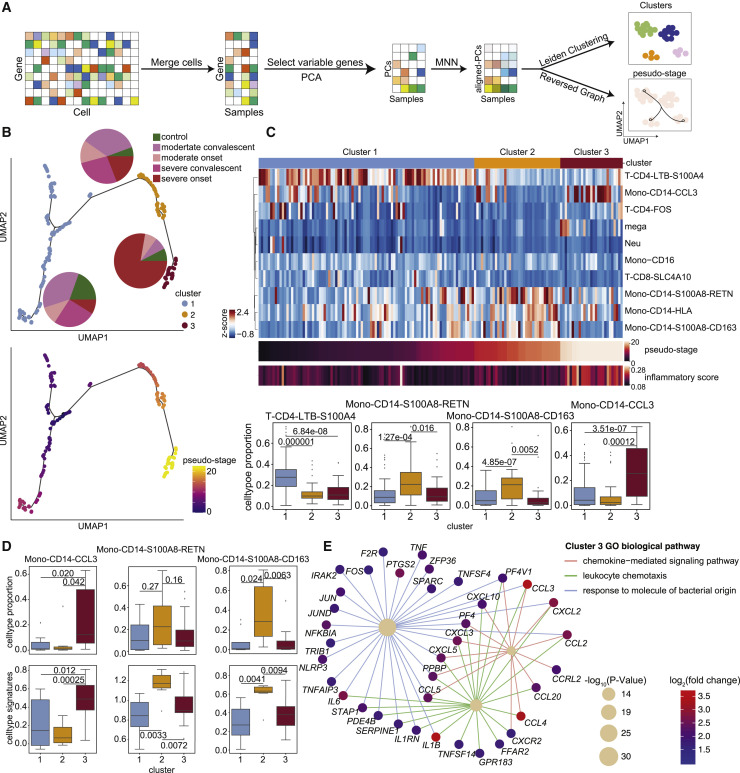
Figure 4.
Heterogeneity analysis of COVID-19 patients illustrates a “three-stage” model associated with inflammatory signatures in monocytes
(A) A workflow schematic for the heterogeneity analysis of COVID-19 patients.
(B) UMAP embeddings of samples from COVID-19 patients, colored by cluster (top) and pseudostage (bottom). Pie plots within the top panel show the proportion of sample states for each cluster.
(C) Illustration of the proportion analysis of the ten hyperinflammatory cell subtypes in COVID-19 patients. Top, heatmap of cell-type proportions for each hyperinflammatory cell subtype in COVID-19 patients. The color bar above the heatmap indicates different clusters. Middle, heatmaps of the ordered pseudostage and corresponding inflammatory score in samples from COVID-19 patients. Bottom, boxplots of the cell proportion of each sample for these four subtypes (from left to right: T-CD4-LTB-S100A4, Mono-CD14-S100A8-RETN, Mono-CD14-S100A8-CD163, Mono-CD14-CCL3) from the indicated clusters (cluster 1, n = 93; cluster 2, n = 37; cluster 3, n = 27).
(D) Top, boxplots of the cell proportions of samples for each subtype (from left to right: Mono-CD14-CCL3, Mono-CD14-S100A8-RETN, Mono-CD14-S100A8-CD163) in different clusters (cluster 1, n = 8; cluster 2, n = 7; cluster 3, n = 21) from severe active patients. Bottom, boxplots of the signature scores of each subtype in different clusters from severe active patients.
(E) Diagram of biological pathways and genes enriched in samples of cluster 3 from severe active patients. Network edges represent gene-pathway associations, and edges of different pathways are indicated by different colors. The significance of the pathways is indicated by circle size. The color bar (from blue to red) represents fold change values in gene expression levels, p values were assessed by clusterProfiler’s built-in function “enrichGO” with default parameters.
In (C) and (D), statistical significance was evaluated with Student’s t test. The boxplots show the mean and interquartile range (IQR), with whiskers extending to 1.5 × IQR.