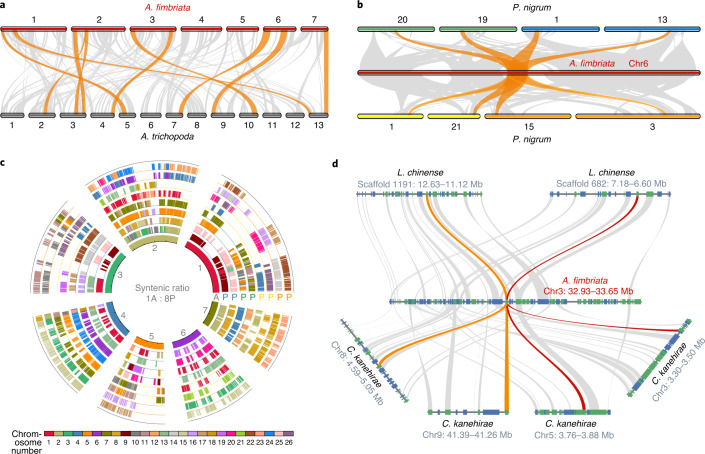
Fig. 2. Intergenomic comparisons revealed that A. fimbriata lacks any WGD after the shared WGD in the common ancestor of all angiosperms and identified two novel WGDs in P. nigrum.
a, Syntenic comparison between A. fimbriata and A. trichopoda37 revealed a 1:1 ratio that suggests no lineage-specific WGD in A. fimbriata after its divergence from A. trichopoda. Syntenic blocks with more than ten genes are linked by grey lines; the largest ten syntenic blocks are highlighted in orange. b, Three rounds of WGDs in P. nigrum were identified via syntenic comparison to A. fimbriata, a finding in contrast to the single WGD in a previous report4. Exemplar syntenic relationships of eight regions in P. nigrum matching a single genomic region in A. fimbriata are highlighted in orange. c, Chromosome-level syntenic alignments of P. nigrum to the A. fimbriata reference genome further support three WGDs. The inner circle represents the seven chromosomes of A. fimbriata (marked with A) and the outer eight circles illustrate the corresponding syntenic regions in P. nigrum (marked with P). The corresponding circles with the same colour of ‘P’ represent synteny from the most recent WGD (Pn-α); the circles where ‘P’ is coloured blue, green, yellow or orange were duplicated in the Pn-β event. d, Microsynteny comparisons clarified the timing of other previously reported WGDs in magnoliids. Representative synteny relationship shows that one A. fimbriata region matches two regions in L. chinense and four regions in C. kanehirae. Rectangles represent annotated genes with orientation on the same strand (blue) or reverse strand (green) and the grey lines connect syntenic gene pairs, with one set highlighted in colour.