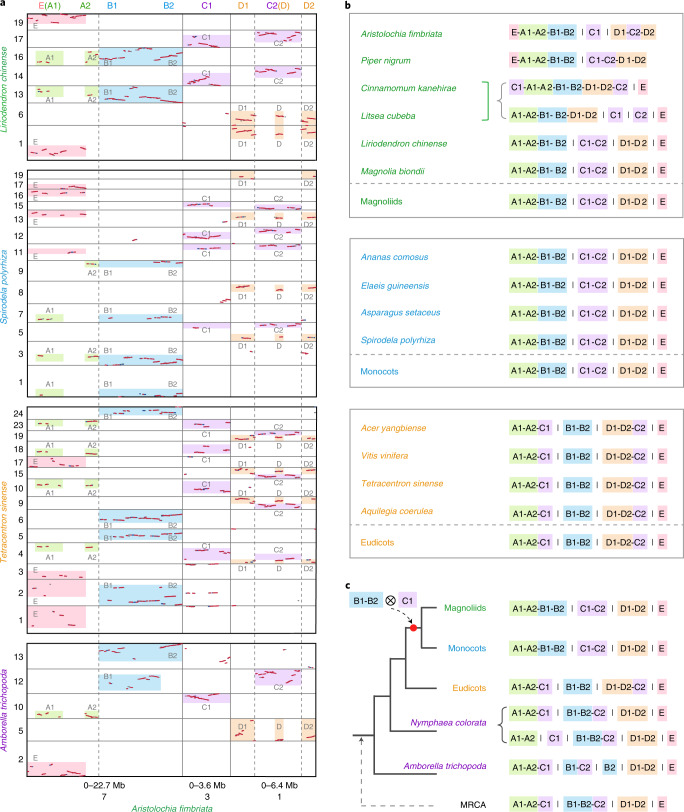
Fig. 3. Common genomic rearrangements present in magnoliids and monocots but absent from eudicots and the two representative species of the ANA grade.
a, The local syntenic blocks identified between the A. fimbriata genome and the genomes of A. trichopoda, T. sinense, S. polyrhiza and L. chinense. The specific genomic regions associated with the Af7 fusion were named regions of E, A1, A2, B1, B2, C1, C2, D, D1 and D2 as marked on top of the plot. The A1 region seems to be embedded in the E region, and is indicated as E(A1). Similarly, the D region seems to be embedded in the C2 region and is indicated as C2(D). Highlighted regions represent syntenic blocks among the compared genomes. The grey dotted lines in a indicate the fusion point in chromosome 7 of A. fimbriata. b, The connection patterns of the orthologous regions in the representative genomes of magnoliids, monocots and eudicots. There are two different connection patterns for the paralogous regions in the C. kanehirae and L. cubeba genomes and both patterns were presented. c, The inferred topology of angiosperms based on a common genomic exchange event shared by monocots and magnoliids. MRCA here represents the most recent common ancestor of extant angiosperms.