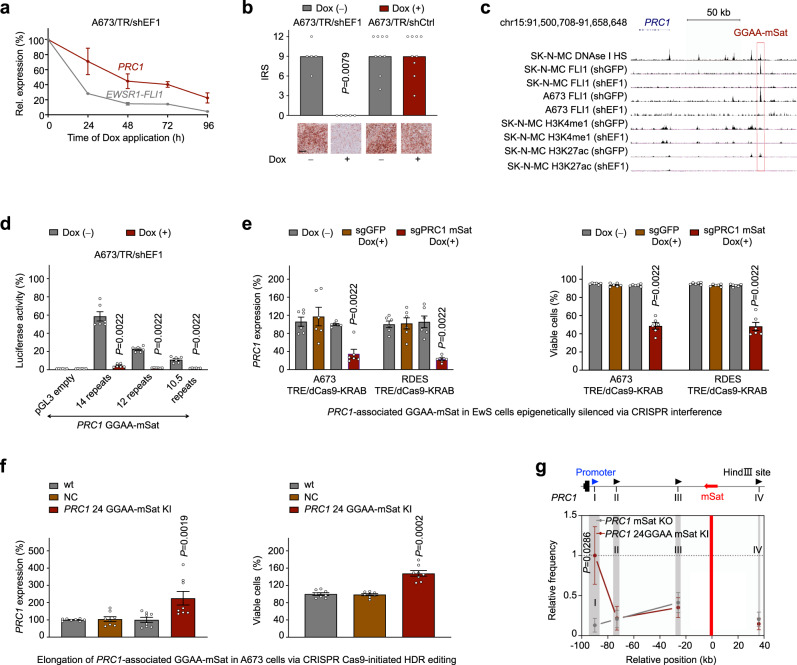
Fig. 2. PRC1 is a direct EWSR1-FLI1 target gene.
a Time-course knockdown of EWSR1-FLI1 in A673/TR/shEF1 EwS cells harboring a Dox-inducible shRNA against EWSR1-FLI1 and analysis of PRC1 and EWSR1-FLI1 expression by qRT-PCR in vitro. Dots represent means and whiskers SEM, n = 4 biologically independent experiments. b Analysis of PRC1 expression by IHC in xenografts derived from A673/TR/shEF1 and A673/TR/shCtrl cells with/without Dox-treatment for 96 h in vivo. Data are displayed as individual dots. Horizontal bars represent the median, n indicates the number of biologically independent experiments/cell line: A673/TR/shCtrl n = 9, A673/TR/shEF1 n = 5; scale bar = 200 µm. c Integrative genomics view (hg19) of the PRC1 locus from data of A673 and SK-N-MC cells being transfected with shRNAs targeting either GFP (shGFP; negative control) or EWSR1-FLI1 (shEF1). d Luciferase reporter assays in A673/TR/shEF1 cells with/without knockdown of EWSR1-FLI1 (Dox+/−) 72 h after transfection. Data are displayed as individual dots. Horizontal bars represent means and whiskers SEM, n = 6 biologically independent experiments. e Analysis of PRC1 expression by qRT-PCR (left) and viable cells (right) in TRE-regulated dCas9-KRAB A673 and RDES cells transduced with sgRNAs against the PRC1-associated GGAA-mSat and GFP sgRNAs (negative control) 15d after addition of Dox (2 µg/ml). Data are displayed as individual dots. Horizontal bars represent means and whiskers SEM, n = 6 biologically independent experiments. f Analysis of PRC1 expression by qRT-PCR (left) and viable cells (right) in wildtype (wt), CRISPR Cas9-edited negative control (NC), and CRISPR Cas9-initiated HDR edited (insertion of 24-GGAA-repeats) A673 cells 72 h after seeding. Data are displayed as individual dots. Horizontal bars represent means and whiskers SEM, n = 8 biologically independent experiments. g Relative crosslinking frequencies observed in CRISPR Cas9-initiated HDR edited PRC1-associated GGAA-mSat knockout (KO) A673 EwS cells are shown in gray and insertion of 24-GGAA-repeats in dark red. Gray shading indicates the position and size of the analyzed fragments, and red shading the ‘vantage point’ fragment harboring the PRC1-associated GGAA-mSat. The highest crosslinking frequency value in the graph was normalized and set to 1 in each replicate. Data are mean and SEM, n = 4 biologically independent experiments. Two-sided Mann-Whitney test in all panels.