Fig. 2. A family of EsGAP venom proteins contributes to escape behaviour.
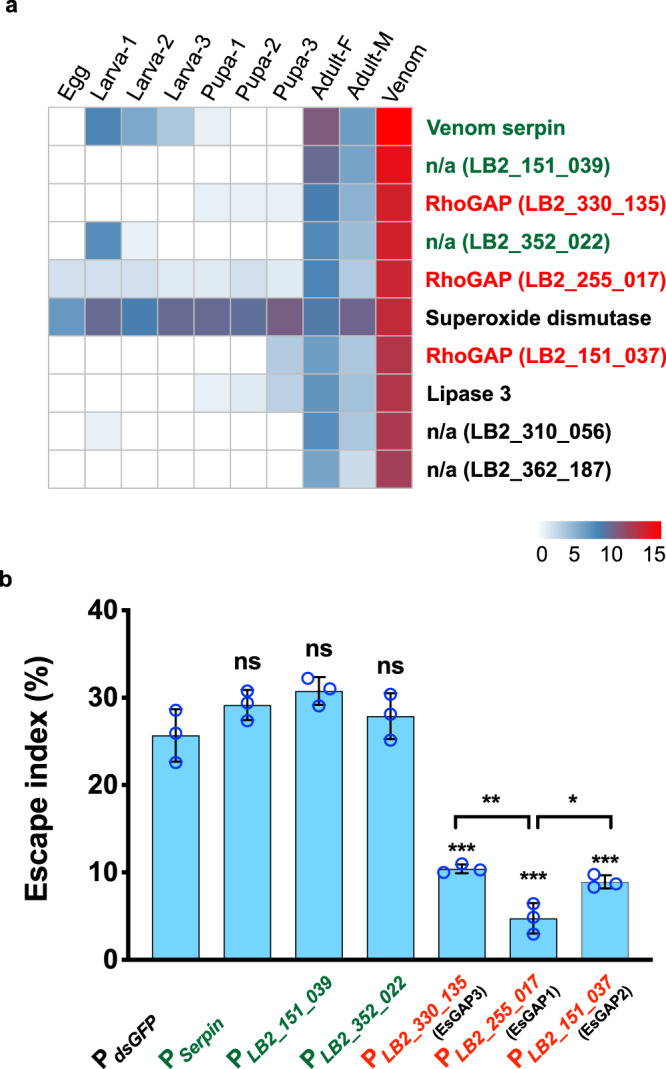
a Transcription heatmap of all venom-protein genes that were significantly highly expressed in venom. Genes are displayed in order of expression levels in the venom. The three genes in red were predicted to have RhoGAP domains and were subjected to functional testing together with another three highly expressed genes (in green). b Escape indices of host larvae that were parasitized by Lb with knockdown of the indicated proteins using an RNAi strategy. dsGFP-treated Lb (PdsGFP) was used as a control. Drosophila larvae infected by parasitoids with knockdown of EsGAP had the lowest escape index. The number of larvae exhibiting escape behaviour was recorded from 30 min to 75 min after exposure to Lb. Left to right: n = 360, 278, 323, 424, 317, 391 and 316 biologically independent host larvae. Three biological replicates were performed. Data are presented as mean values ± SD. Significance was determined by two-sided unpaired Student’s t test (PSerpin: P = 0.1564; PLB2_151_039: P = 0.0609; PLB2_352_022: P = 0.3953; PEsGAP3: P = 0.001; PEsGAP1: P = 0.0005; PEsGAP2: P = 0.0007; PEsGAP3 vs PEsGAP1: P = 0.0056; PEsGAP2 vs PEsGAP1: P = 0.0188; *P < 0.05; **P < 0.01; ***P < 0.001; ns: not significant). Source data are provided as a Source data file.
