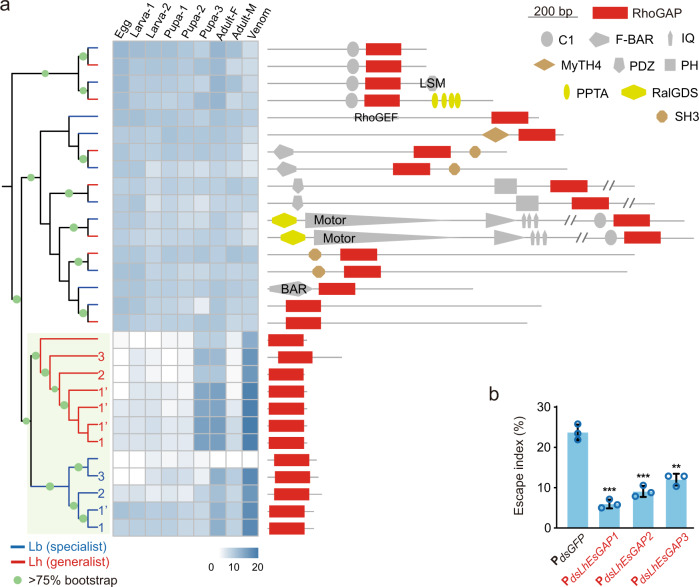
Fig. 5. Molecular evolution of RhoGAP-containing genes in Leptopilina.
a Phylogenetic relationship across all potential RhoGAP domains encoded in Lh and Lb genomes. Branches being followed with a number indicate the corresponding termed EsGAP genes with functional evidence. Note that there are tandem duplicated copies of EsGAP1 in both Lb and Lh, which share the near-identical sequence with EsGAP1 and therefore are termed as LbEsGAP1’ and LhEsGAP1’, respectively. The expression profile of each locus is shown in the heatmap. The actual gene length is indicated by the line and schematic gene architecture is shown with annotated domains. See detailed information in Supplementary Table 3. b Escape indices of host (D. melanogaster) that were parasitized by dsRNA-treated Lh. LhEsGAP1, LhOGS01638; LhEsGAP2, LhOGS20221; LhEsGAP3, LhOGS01640. dsGFP-treated Lb (PdsGFP) was used as a control. The number of larvae exhibiting escape behaviour was recorded from 30 min to 75 min after exposure to Lh. Left to right: n = 424, 357, 341 and 355 biologically independent host larvae. Three biological replicates were performed. Data are presented as mean values ± SD. Significance was determined by two-sided unpaired Student’s t test (PdsLhEsGAP1: P = 0.0002; PdsLhEsGAP2: P = 0.0004; PdsLhEsGAP3: P = 0.0011; **P < 0.01; ***P < 0.001). Source data are provided as a Source data file.