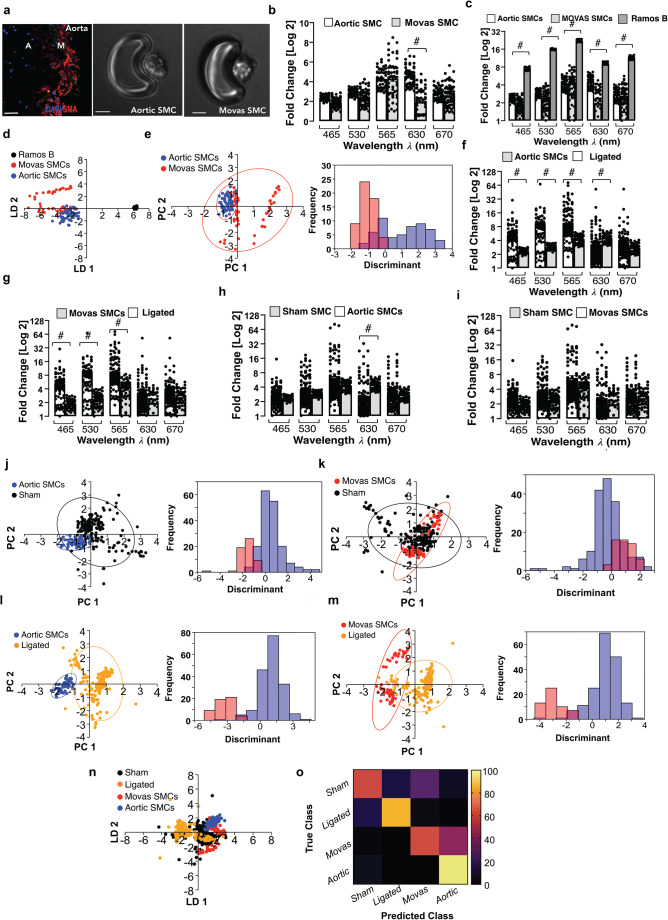
Fig. 2.
Single cell photonics of aortic SMCs ex vivo and cultures aortic Movas SMCs in vitro. (a). Representative immunohistochemical analysis of ⍺-actin (SMA) expression in mouse aorta and visualisation of aortic SMCs and Movas SMCs on each V-cup in the LoaD platform. (b). Single cell photon emissions across five wavelengths from freshly isolated aortic SMCs and cultured aortic Movas SMCs in vitro. Data are the Log2 fold increase and represent the mean ± SEM of 55 cells/group, #p≤0.001. (c). Single cell photon emissions of aortic and Movas SMCs compared to Ramos B cells. Data are the Log2 fold increase and represent the mean ± SEM of 55 cells/group, #p≤0.001. (d). LDA plots of aortic SMC (blue), Movas SMC (red) and Ramos B (black) cells in vitro. Data are from 165 cells across the five wavelengths (e). PCA loading and LDA plots of aortic SMC and Movas SMC. (f-i). Single cell photon emissions from (F, H) aortic SMC and (G, I) Movas SMC cells compared to sham (F, G) and ligated cells (H, I) ex vivo. Data are the Log2 fold increase and represent the mean ± SEM of 55–178 cells/group, #p≤0.001 vs Sham (F,G) Ligated (H,I). (j-m). PCA loading plots and LDA of aortic SMCs (J, L) and Movas SMCs (K, M) compared to carotid artery SMCs (sham) (J) and ligated cells, respectively. (n). LDA plots of sham (black), ligated (orange) cells ex vivo compared to aortic SMCs (blue) and Movas SMC (Red) cells in vitro G. Confusion matrix of true class and predicted class following a leave-one-outcross-validation procedure by the LDA classifier. Data are from 466 cells across five wavelengths