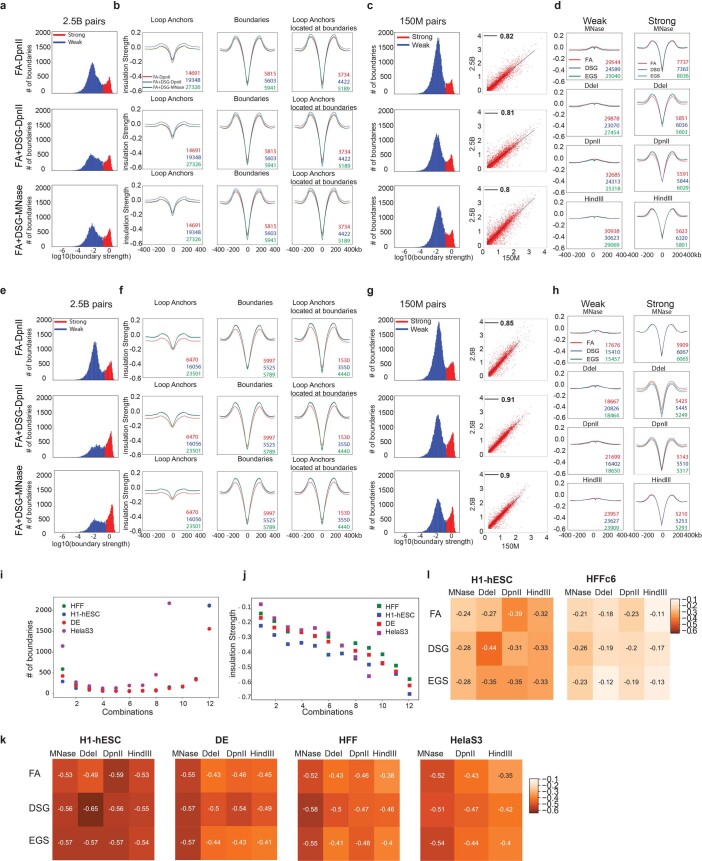
Extended Data Fig. 7. Insulation quantification is robust to experimental variations.
a-d. Rows: HFFc6 deep data from FA-DpnII (top), FA+DSG-DpnII (middle) and FA+DSG-MNase (bottom). Columns: boundary strength distributions with strength threshold (a) (Methods), pileups in FA-DpnII, FA+DSG-DpnII and FA+DSG-MNase for aggregate insulation scores at loop anchors (left), strong insulation boundaries (middle) and loop anchors colocalizing with strong insulation boundaries (right) (b). Left panel: boundary strength distribution of matrix data (Fig. 1a) for FA-DpnII, FA+DSG-DpnII and FA+DSG-MNase. Right panel:correlation of strong boundaries between deep and matrix data for FA-DpnII, FA+DSG-DpnII and FA+DSG-MNase (c). Aggregate insulation profile of weak and strong boundaries from matrix data for HFFc6 for cross-linkers and nucleases (d). e–h. H1-hESC data displayed like Extended Data Fig. 7a-d. i. Number of boundaries (y-axis) stratified by number of protocols (1 to 12; see Fig. 1a) wherein a given boundary was detected (x-axis). j. Insulation strength of boundaries stratified as in (i). k. Mean insulation strength for boundaries detected in at least half of protocols for various cross-linkers and enzyme combinations of H1-hESC, DE, HFF and HeLa-S3-NS (Methods). l. Mean insulation strength of loop anchors detected in all three deep protocols for both HFFc6 and H1-hESC, averaged for 12 protocols of H1-hESC and HFF.