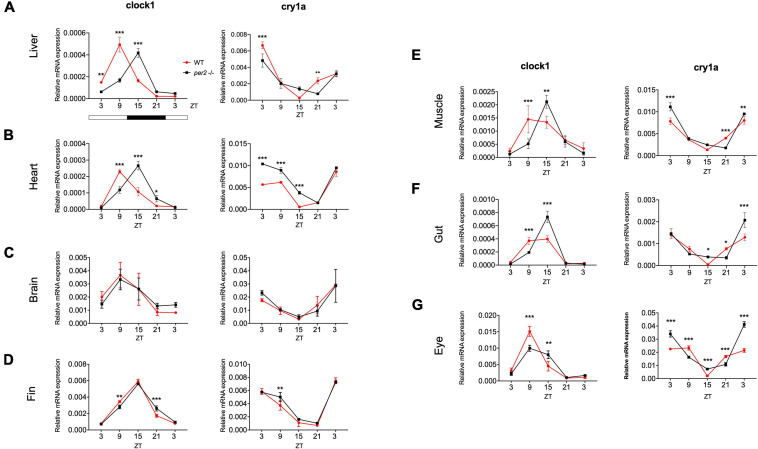
FIGURE 4.
Per2 knockout alters rhythmic mRNA expression of the cry1a and clock1 clock genes in adult zebrafish tissues. qRT-PCR analysis of expression levels of the circadian clock genes clock1 and cry1a in the (A) liver, (B) heart, (C) brain, (D) fin, (E) muscle, (F) gut and (G) eyes of WT and per2 mutant adult fish. Sets (n = 3) of 4 WT and per2 KO fishes (each set containing two males and two females) were maintained under LD cycle (14 h light-10 h dark) conditions, tissues were dissected and pooled for RNA extraction at 6-h intervals during a sampling window of 24 h. Mean mRNA relative expression ± SD is plotted on the y-axis; zeitgeber time (ZT) is plotted on the x-axis. Zeitgeber times are indicated for each sample. ZT0 corresponds to lights-on, ZT14 to lights-off. A change in rhythm was observed for both genes in liver, heart, fin, muscle, gut, and eyes, as determined by a significant ANOVA Genotype × Time interaction effect (p < 0.001, see Table 1). For both genes, no effect was observed in the brain. Asterisks represent levels of significance in comparing the two genotypes at each time point individually, corrected by Sidak’s method (***p < 0.001, **p < 0.01, *p < 0.05).