FIGURE 8.
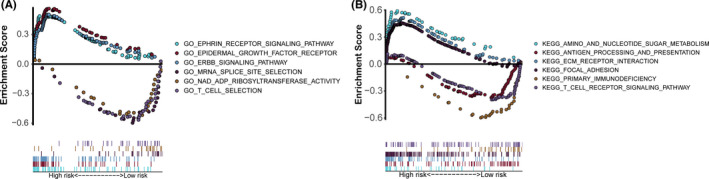
Gene set enrichment analysis on the differentially expressed genes between the high‐risk and low‐risk groups. (A) Gene Ontology (GO) enrichment analysis indicated that the genes were enriched in the ephrin receptor signaling pathway, epidermal growth factor receptor signaling pathway, ERBB signaling pathway, mRNA splice site selection, DNA ADP ribosyltransferase activity, and T cell selection. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis showed that these genes were involved in amino sugar and nucleotide sugar metabolism, antigen processing and presentation, extracellular matrix receptor interaction, focal adhesion, primary immunodeficiency, and T cell receptor signaling pathway
