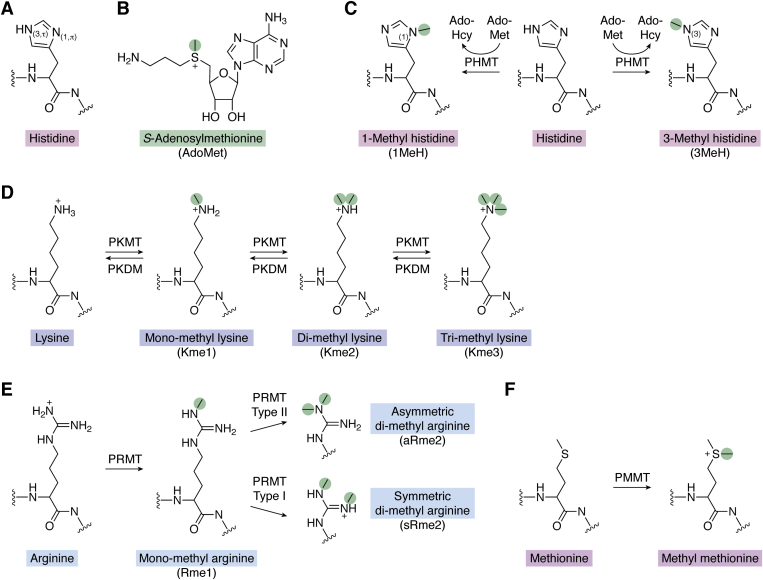
Figure 1.
Biochemistry of protein histidine methylation and selected key cellular protein methylation events.A, histidine structure and nomenclature. The imidazole ring in the histidine side chain contains two nitrogen atoms denoted as N1 (or τ) and N3 (or π). B, chemical structure of AdoMet. The polarized methyl group of AdoMet is highlighted (green circle), and the positive charge of the associated sulfonium ion is indicated. C, biochemistry of histidine methylation. The N1 or N3 atom of the histidine side chain can accept methyl groups through AdoMet-dependent protein histidine methyltransferase (PHMT)-mediated methylation to generate with 1-methylhistidine (1MeH) or 3-methylhistidine (3MeH) as well as the reaction byproduct AdoHcy. D, biochemistry of lysine methylation. Protein lysine methyltransferases (PKMT) can introduce up to three methyl groups to the ε-amino group in the side chain of lysine yielding monomethyl, dimethyl, or trimethyl lysine. In turn, the methyl groups can be removed by protein lysine demethylase (PKDM) enzymes. E, biochemistry of arginine methylation. Protein arginine methyltransferases (PRMT) catalyze the formation of monomethyl arginine. Subsequently, type I and type II PRMTs can catalyze symmetric dimethyl arginine and asymmetric dimethyl arginine, respectively. F, biochemistry of methionine methylation. Protein methionine methyltransferase (PMMT) enzymes catalyze the formation of methyl-methionine from methionine.