TABLE 1.
Sanger sequencing of SARS‐CoV‐2 Spike genes: primers used and RT‐PCR conditions
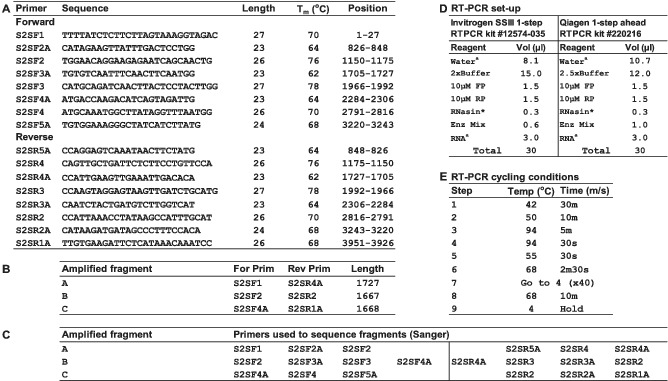
Not e: (A) Primer name, sequence, length, hybridisation temperature (Tm), and location across the SARS‐CoV‐2 genome fragment spanning the Spike glycoprotein open‐reading frame (ORF) are shown. Primers S2SF1 and S2SR1A are positioned in highly conserved regions that flank the S‐gene. Based on the primer position numbers indicated in panel A, residues 94–96 would represent the ATG start codon of the S‐gene. (B) The Spike ORF was amplified in three overlapping fragments using the primers indicated. (C) Sanger sequencing of individual fragments used the primer sets indicated. (D) RT‐PCR set‐up is shown for 1‐step kits supplied by Invitrogen or Qiagen, standard reaction set‐up is shown but awater and aRNA volumes can be adjusted to allow addition of more RNA from specimens yielding rtRTPCR Ct values of ≥25. * Promega RNasin® ribonuclease inhibitor (#N2515). (E) Thermal cycling conditions used on a Bio‐Rad DNA ENGINE DYAD Peltier thermal cycler are shown, with temperatures being calculated.
