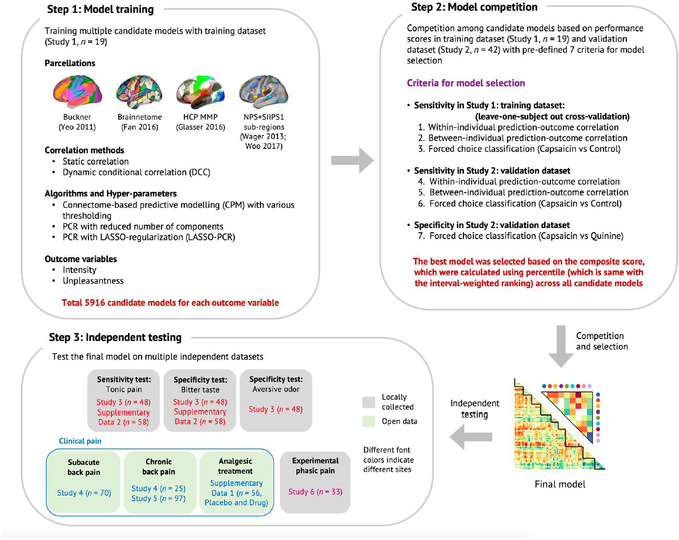
Extended Data Fig. 2 |. Overview of the signature development and test procedure.
Step 1: Model training. Using Study 1 data, we trained a large number of candidate models for each outcome variable (pain intensity and unpleasantness) with multiple combinations of different parcellation solutions (features), connectivity estimation methods (feature engineering), algorithms, and hyperparameters. We generated a total of 5916 candidate models for each outcome variable. Step 2: Model competition. To select the best model, we conducted a competition among the candidate models based on the pre-defined criteria including sensitivity, specificity, and generalizability based on cross-validated performance in Study 1 and also using Study 2 data as a validation dataset. For the full description of the competition procedure, please see Methods, and for the full report of the competition results, please see Extended Data Fig. 3. Step 3: Independent testing. To further characterize the final model in multiple test contexts, we tested the final model on multiple independent datasets, including two additional tonic pain dataset (Study 3 and Supplementary Data 2), three clinical pain datasets (Studies 4–5 and Supplementary Data 1), and one experimental phasic pain dataset (Study 6). Gray boxes represent locally collected datasets, and green boxes represent publicly available datasets. Different font colors indicate different scan sites.