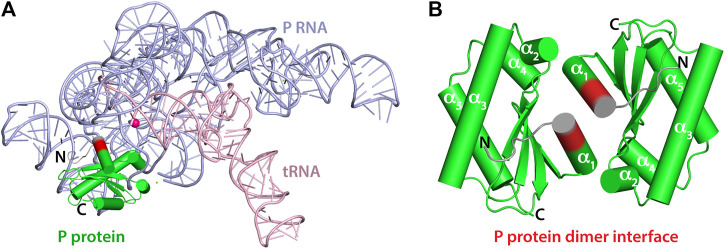
FIGURE 1.
Crystal structures of T. maritima RNase P components. (A) Crystal structure of the holoenzyme in complex with tRNA (PDB 3Q1Q). RNase P RNA component is shown in light blue, the P protein component in green, and tRNA in light pink. The enzyme active site is denoted by the location of a catalytic magnesium ion (magenta sphere). RNAs are represented as loops (backbones) and sticks (nucleobases) and the P protein is represented as cylinders (α-helix) and arrows (β-sheets). Protein residues 14–17 are highlighted in red to indicate the location of the dimerization interface of the P protein alone (PDB 1NZ0). Residues 8–21 play a critical role in binding the P RNA as part of the holoenzyme complex and in optimally aligning the 5’ leader ptRNA substrate. (B) 1.2 Å resolution crystal structure of the P protein shown as a dimer, with molecules A and C (PDB 1NZ0). Protein residues 14–17 are highlighted in red and the N-termini in colored grey. Residues 1–15 are oriented differently between A and B.