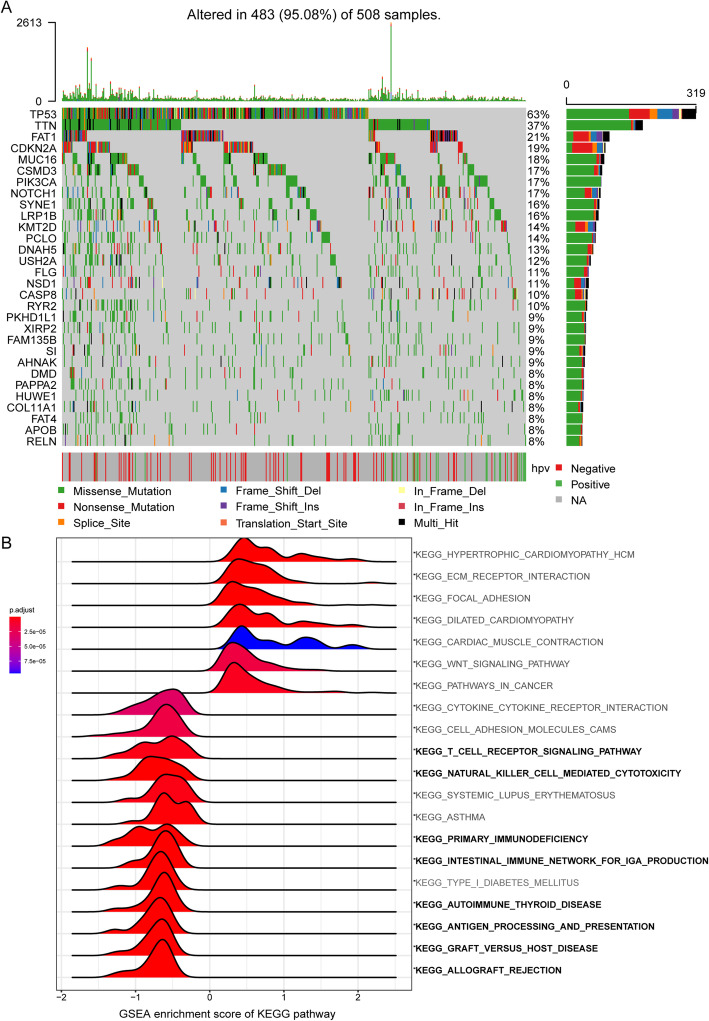
Fig. 1.
TP53 mutation-associated genes in the TCGA HNSC cohort and GSEA of KEGG pathways. A The mutated genes were listed according to their mutation frequencies, considering the status of HPV infection. The mutations include missense mutations, nonsense mutations, splice-site mutations, frameshift mutations, and in-frame mutations. The oncoplot were drawn in maftools package. B Gene set enrichment analysis (GSEA) was performed to enrich the KEGG pathways in genes that were related to TP53 mutation. Significant enrichments in immune-related KEGG pathways are labeled in bold. A false discovery rate (FDR) of less than 0.05 and an absolute value of the enrichment score (ES) of greater than 0.5 were defined as the cutoff criteria. The analysis were finished using clusterProfiler package