Figure EV5. Missing values and batch effects in the Aging mouse study.
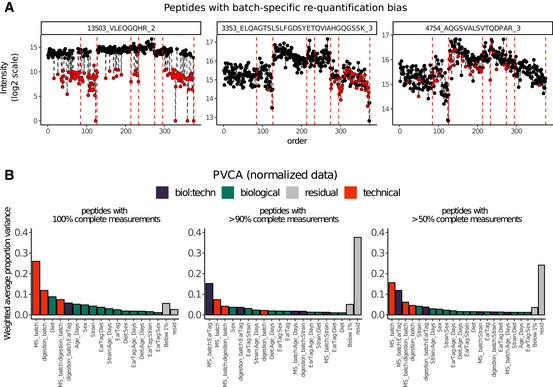
(A) Re‐quantification of elution traces can pick up batch‐specific noise that can be drastically different (left panel) or indistinguishable (middle and right panel) from confidently identified peptide fragments, meaning that values inferred should be treated with extreme caution. Black points are regular quantifications, red points are re‐quantifications; (B) PVCA can only be applied to complete matrices, and thus, missing values need to be inferred. This makes this method highly sensitive to missing values inference (here filled with 0). Depending on the completeness cutoff, variance distribution across technical and biological factors varies substantially. Note that when peptides with missing values are used in the analysis (panels center and right), a substantial portion of variance is attributed to "resid" (residual), meaning that this variance cannot be associated with any of known factors, indicating that missing value distribution is at least partially random.
