Figure 7. PIN2 dephosphorylation at S439 is important for modulation of primary root growth and lateral root density in response to nitrate treatments.
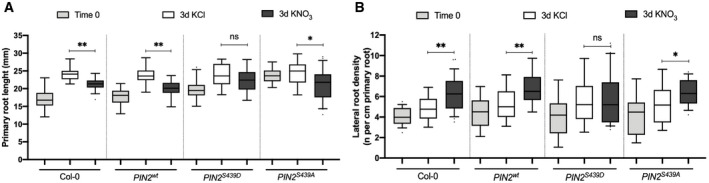
- Primary root length of the different genotypes was measured using the ImageJ program 3 days after 5 mM KNO3 or KCl treatments. Tukey box plot show results from 3 independent biological replicates per experimental condition (n = 8–10 roots each replicate). The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5x IQR (interquartile range). Outlier data are plotted individually. Asterisk indicates statistically significant difference between means analyzed by unpaired, two‐tailed and assuming equal variance t‐test (*P < 0.05, **P < 0.01).
- Number of initiating and emerging lateral roots for all genotypes were counted using light microscopy 3 days after 5 mM KNO3 or KCl treatments. Tukey box plot show results from 3 independent biological replicates per experimental condition (n = 8–10 roots each replicate). The box plot shows the data within the interquartile range (25th and 75th percentiles) and a solid black line represents the median. Whiskers show maximum and minimum values no further than 1.5× IQR (interquartile range). Outlier data are plotted individually. Asterisk indicates statistically significant difference between means analyzed by unpaired, two‐tailed and assuming equal variance t‐test (*P < 0.05, **P < 0.01).
Source data are available online for this figure.
