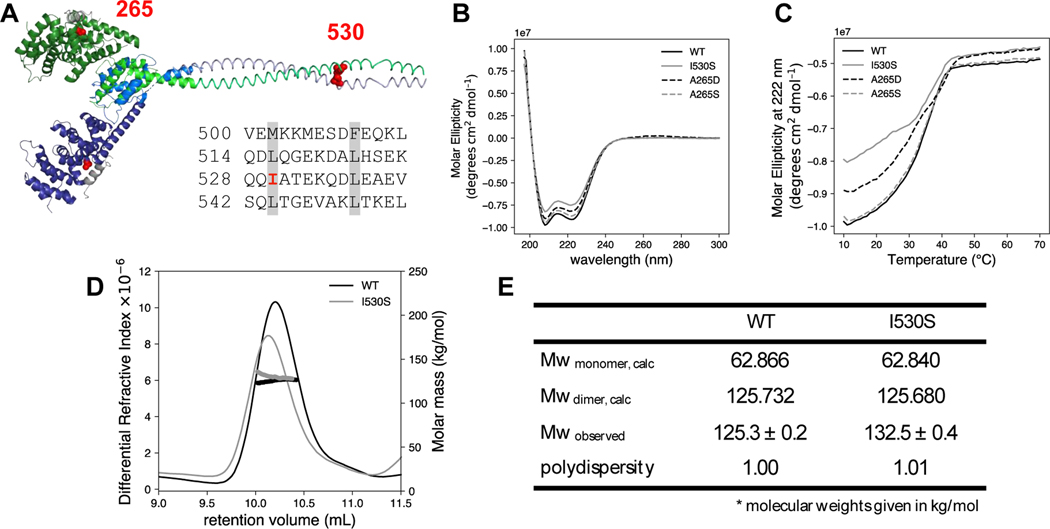
Figure 3. Structural characterization of NT mutants.
(A) The predicted DID and dimerization domain structure of DIAPH1 were constructed from the PDB files 2BNX and 2F31 and from the prediction program COILS 43. The DD sequence is shown with the heptad repeat of hydrophobic residues highlighted in gray. The two DID-DD chains are each displayed in shades of blue and green. The DAD helices are shown in gray, and mutated residues 265 and 530 are in red spheres. Circular dichroism (B) wavelength scan and (C) thermal denaturation monitored at 222 nm for DIAPH1-NT WT and mutants. Conditions: 3 μM protein in phosphate-buffered saline. SEC-MALS analysis was done on both WT and the I530S mutant of DIAPH1. (D) Plot of differential refractive index and molecular mass (thick line) vs elution volume. The observed average molecular weights in panel (E) are closest to the dimer calculated Mw. The SEC-MALS data is representative of two trials, with separate protein preparations. In the other trial, Mw/polydispersity: WT, 133.7/1.04; I530S, 129.6/1.18.