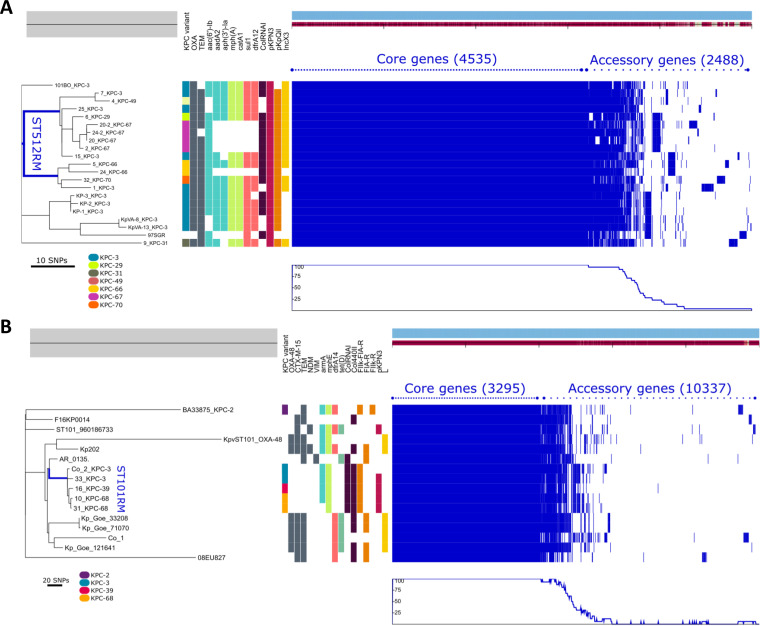
FIG 3.
Phylogenetic analysis of ST512 (A) and ST101 (B) K. pneumoniae lineages. The left side shows the unrooted maximum likelihood phylogenetic trees based on a concatenated core gene alignment. The middle represents metadata, each block indicating the presence or absence of a specific resistance gene and of the plasmid replicon (some of which are not shown for the sake of clarity). A legend shows the colors assigned to each KPC variant in the respective column. The right side shows a gene possession matrix, with each row representing the respective strain gene content. Each of the blue blocks on the right represents the presence of a gene. (A) Phylogenetic comparison of ST512 genomes performed using 10 CZA-resistant and 3 CZA-susceptible strains, isolated in the same surveillance period in the hospital and on 7 genomes of epidemiologically unrelated ST512 strains. Among them, four genomes were downloaded from the collection of ST512 strains at the Klebsiella Pasteur MLST isolate database (https://bigsdb.web.pasteur.fr/klebsiella), and three were KPC-3 ST512 producers isolated at the University Hospital Policlinico Umberto I of Rome in 2012 (KP-1, KP-2, KP-3). The blue line localizes the branches of the tree generated by genomes of the CZA-resistant strains under study (except strain 9 producing KPC-31, which was located in a different branch). (B) Phylogenesis of ST101 strains performed on three CZA-resistant strains (strains 16, 10, and 31) and one CZA-susceptible strain (strain 33) isolated during the surveillance period and for comparison on 11 ST101 genomes downloaded from the NCBI GenBank, including two ST101 strains isolated from patients hospitalized in the ICU ward dedicated to COVID-19 patients and identified in our hospital in April 2020 (strain Co_1, producing OXA-48, and strain Co_2, producing KPC-3).