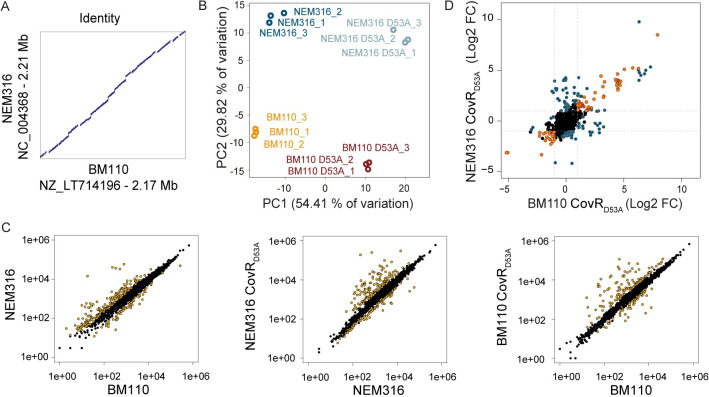
Fig 5. Plasticity of the CovR regulon in BM110 and NEM316.
(A) Sequence identity between the NEM316 and BM110 chromosomes. The two chromosomes are co-linear with vertical and horizontal gaps corresponding to strain-specific sequences, usually due to prophages and ICE. (B) Principal component analysis of RNA-seq data for the BM110 and NEM316 WT strains and their corresponding CovRD53A mutants. Biological triplicates are resolved by PCA with variability sustained by CovR inactivation (PC1) and WT differences (PC2). (C) Scatter plots of the normalized read counts for each of the 1,716 homologous genes in the two WT strains, and between the WTs and their corresponding CovRD53A mutants. Yellow dots represent significant differential transcription (adjusted p-value < 0.005). (D) Pairwise comparison of the fold change in the two CovRD53A mutants. Genes with a similar and significant fold change upon CovR inactivation in the two backgrounds are symbolized with orange dots. Genes with a significant fold change in one mutant only or which show a significant different fold change in the two mutants are highlighted with blue dots.