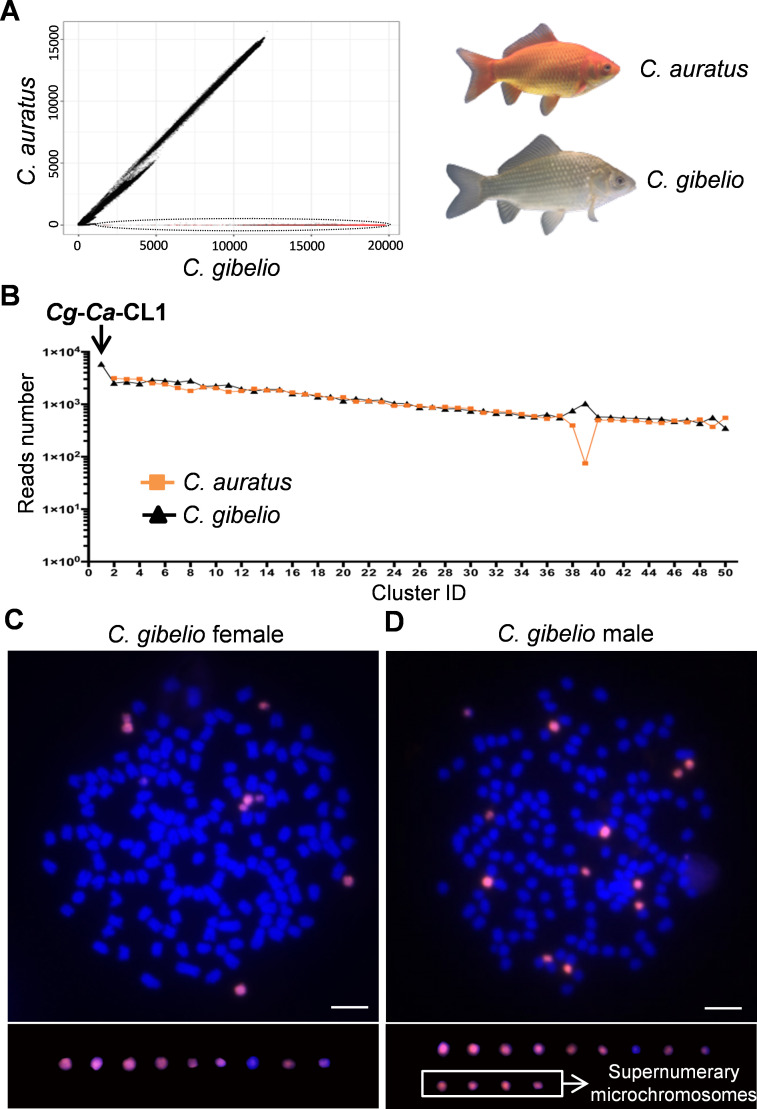
Fig 1. Repetitive sequence expansion on microchromosomes.
(A) Pairwise comparison of all analyzed reads between hexaploid C. gibelio and tetraploid C. auratus. The X-axis and Y-axis show the numbers of similarity hits for each read in C. gibelio and C. auratus, respectively. Each spot corresponds to one read. The black ellipse indicates the repetitive sequences with expansion in C. gibelio compared to C. auratus. The red dots represent the reads of the most expanded satellite cluster (Cg-Ca-CL1). (B) The top 50 largest repeat clusters generated by C. gibelio-C. auratus (Cg-Ca) pairwise comparative analysis. The Y-axis shows the reads number in clusters, and the X-axis shows the cluster ID. (C, D) FISH analysis of satellite repeat cluster Cg-Ca-CL1 in female metaphase (C) and male metaphase (D) of C. gibelio. Scale bar = 5 μm. The white square indicates supernumerary microchromosomes with a male determination role in C. gibelio.