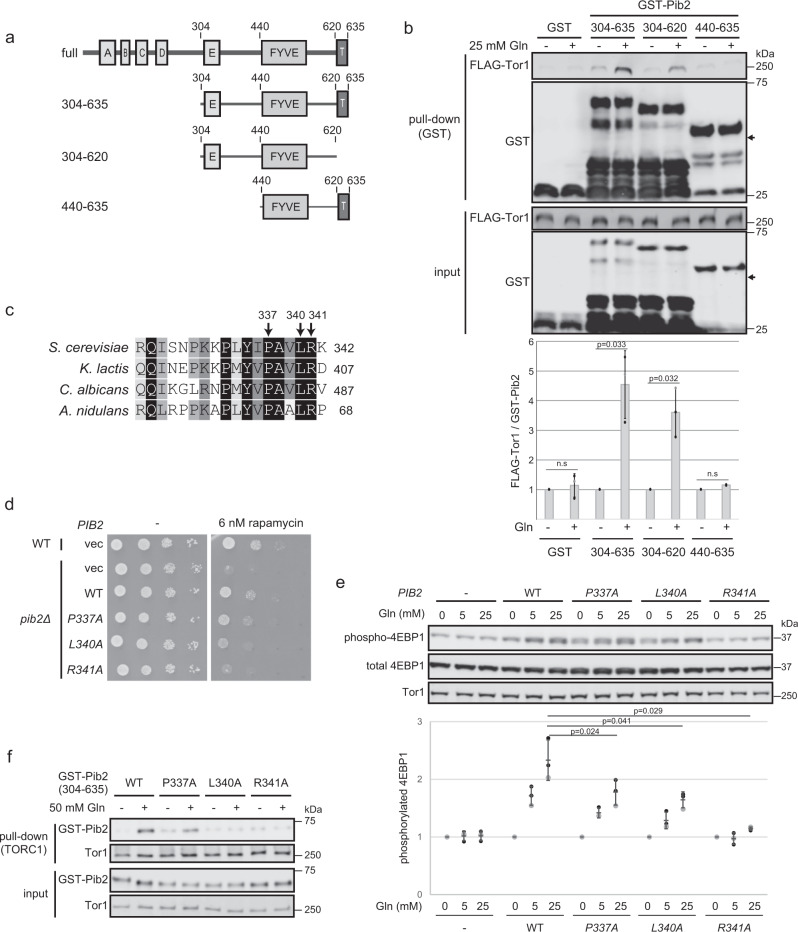
Fig. 2. The Pib2 E motif is required for its glutamine-induced interaction with TORC1.
a Schematic diagram of full-length Pib2 and the deletion mutants is used in (b). b The E-motif containing the central region of Pib2(304-439) is required for the glutamine-induced interaction between Pib2 and TORC1. Pull-down assays were performed using yeast cell lysates and bacterially-expressed GST-Pib2 deletion mutants (as in Fig. 1b). The bar graph shows GST-Pib2-bound FLAG-Tor1 normalized to the samples without glutamine. Error bars represent the standard deviation (n = 3 independent experiments), and significance was determined by a two-tailed Student’s t-test. c The E motif in Pib2 is conserved among species. Sequence alignment of the E motifs in Pib2-like proteins from the indicated species. d Pib2 E motif mutants display rapamycin sensitivity. Wild-type (TM141) or pib2Δ (MH1059) strains were transformed with a vector (pRS416) or plasmids encoding wild-type PIB2 (pMH342) or pib2 mutants (pMH497, pMH499, or pMH498). Serially diluted cell suspensions were spotted on SD plates lacking uracil with or without 6 nM rapamycin and grown at 30 °C for 3 days. e Pib2 E motif mutants decrease TORC1 activity in vitro. An in vitro TORC1 kinase assay was performed (as for Fig. 1d) with permeabilized yeast cells prepared from pib2Δ (MH1059) cells carrying a vector (p416ADH) or plasmids encoding wild-type PIB2 (pMH330) or pib2 mutants (pMH510, pMH512, or pMH514). Dot plots represent the ratio of phosphorylated/total 4EBP1 normalized to the samples without glutamine (n = 3 independent experiments). f Pib2 E motif mutants are deficient for the glutamine-induced interaction with TORC1. Purified TORC1 on magnetic beads was incubated with bacterially-expressed GST-Pib2(304-635) mutants with or without 30 mM glutamine. GST-Pib2 co-precipitation with TORC1 was detected by western blotting.