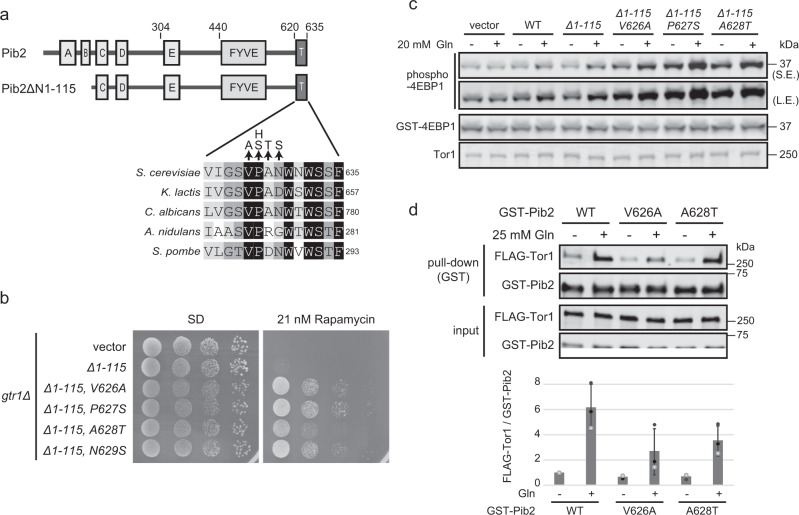
Fig. 3. Active PIB2 mutations are clustered in the conserved tail motif.
a Schematic diagram of wild-type Pib2 and Pib2(Δ1−115) was used for active mutant screening. Mutations in the active mutants from the tail motif are shown with multiple sequence alignments for different species. b Tail motif mutations confer yeast cells with rapamycin resistance. The gtr1Δ strain (MH1030) was transformed with a vector (p416ADH) or plasmids encoding PIB2 mutants (pMH334, pKY11-1, pKY14-1, pKY24-1, or pKY21). Serially diluted cell suspensions were spotted on SD plates lacking uracil with or without 21 nM rapamycin and then grown at 30 °C for 3 (left) or 5 (right) days, respectively. c Pib2 active mutants enhance TORC1 activity in vitro. An in vitro TORC1 kinase assay was performed (as for Fig. 1d) using permeabilized yeast cells prepared from pib2Δ (MH1059) cells carrying a vector (p416ADH) or plasmids encoding wild-type PIB2 (pMH330) or PIB2 mutants (pMH334, pKY11-1, pKY14-1, pKY24-1, or pKY21). L.E long exposure, S.E short exposure. d The interaction between active Pib2 mutants and TORC1 is glutamine-responsive. Pull-down assays were performed using yeast cell lysates and bacterially-expressed GST-Pib2 tail motif mutants (as in Fig. 1b). The bar graph shows GST-Pib2-bound FLAG-Tor1 normalized to the wild-type Pib2 samples without glutamine. Error bars represent the standard deviation (n = 3 independent experiments).