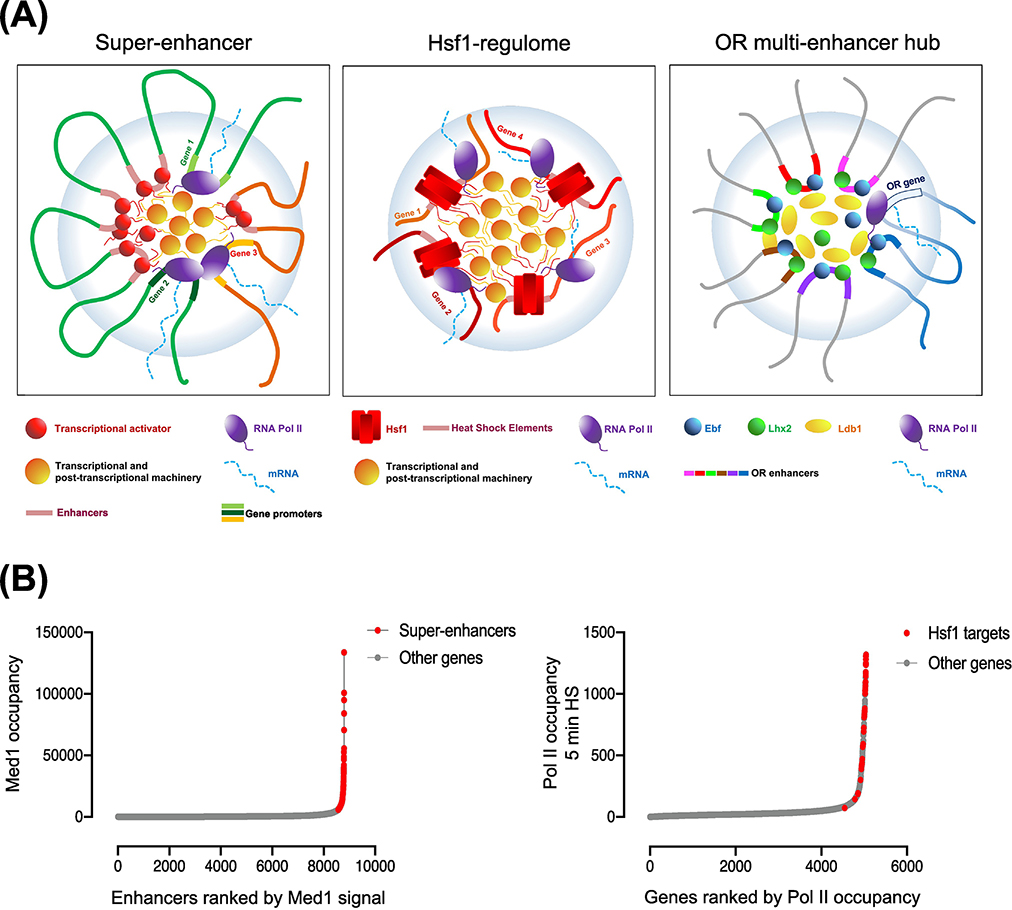
Figure 4. HSP Gene Coalescence Shares Key Attributes with Super-Enhancers and Olfactory Receptor Multi-Enhancer Hubs.
A) (Key Figure)(Left) Super-enhancers (SEs) comprise clusters of enhancers (red spheres) that engage in physical interactions with promoters of associated genes (genes 1–3). SEs are densely occupied by transcriptional activators (red spheres) and other transcriptional and post-transcriptional apparatus (orange ovals), and a network of multivalent cooperative interactions exist between them. The increase in density and affinity of interactions between these components contribute towards SE assembly within a phase-separated condensate (light blue bubble). (Center) Yeast HSP genes (orange) are occupied by unusually high densities of activator (Hsf1, red rectangles), RNA Pol II (purple ovals) and other transcriptional and post-transcriptional factors (orange ovals), and exhibit strong inter-chromosomal interactions upon their induction. (Right) Inter-chromosomal clustering of multiple olfactory receptor (OR) enhancers lead to the formation of an OR enhancer hub in mouse olfactory sensory neurons. The OR enhancers, co-bound by transcription factors (Ebf and Lhx2) and coactivators (Ldb1), stochastically converge on a single olfactory receptor gene to activate its transcription.
B)(Left) Mediator (MED1) occupancy across 8794 enhancers in mouse ESC cells arranged according to their rank order [13]. (Right) Pol II occupancy across 5047 genes in S. cerevisiae in the 5 min heat shock condition, arranged according to their rank order (data from [61]). Both SEs and Hsf1-regulated genes fall above inflection points of their respective curves, suggesting exceptionally high occupancy levels of transcriptional machinery at these genes.