Figure 1.
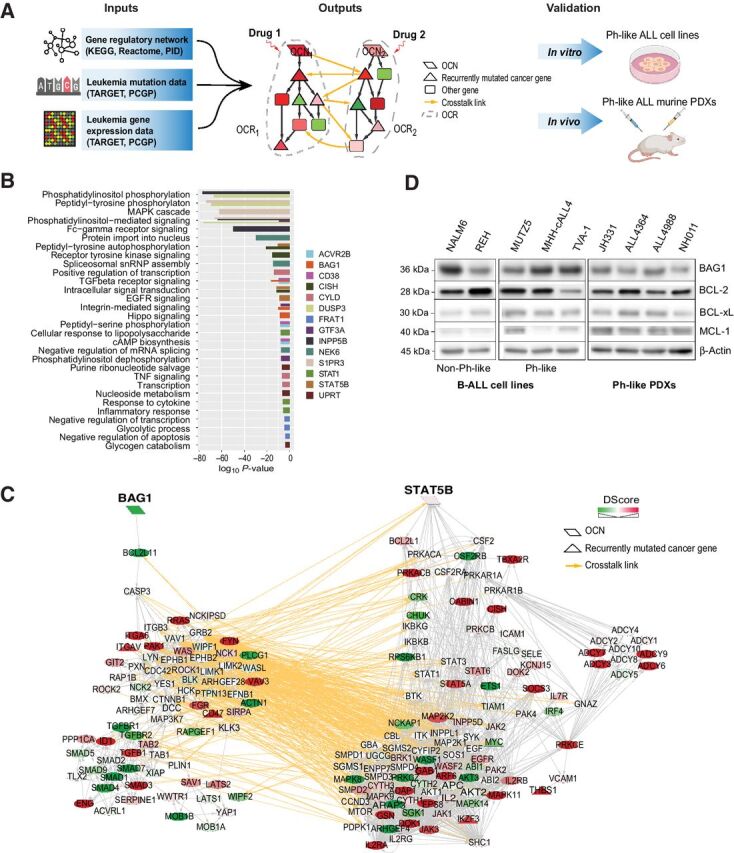
A systems biology approach to discovery and testing of synergistic therapeutic targets. A, Overview of the Optimal Control (OptiCon) network-based approach toward identifying and validating synergistic drug targets in Ph-like B-ALL. OptiCon input data include a high-quality human gene regulatory network integrating expert-curated pathway annotations from three public pathway databases (KEGG, Reactome, and NCI-Nature Pathway Interaction Database), patient genetic mutation data, and gene expression data from TARGET and PCGP consortia (details in Materials and Methods). Output of OptiCon identifies synergistic OCN pairs. Druggable pathways defined by OCNs and their respective OCRs are then validated in vitro in Ph-like ALL cell lines and in vivo in murine PDX models. B, Enriched gene ontology (GO) terms among predicted OCRs. Each color represents the OCR of a predicted OCN. Thickness of bars varies since certain terms were enriched among multiple OCRs. C, Synergistic OCN pair STAT5B and BAG1 predicted for Ph-like ALL. Gene crosstalk links between their specific OCRs are shown in yellow. Shade of a node represents the deregulation score (DScore) of the corresponding gene. Red, up-regulated in Ph-like ALL; green, down-regulated in Ph-like ALL. D, Baseline protein expression of BAG1 and the anti-apoptotic BCL2-family proteins BCL2, BCL-xL, and MCL1 in several B-ALL and Ph-like ALL cell lines and Ph-like PDXs.
