Abstract
Body weight at the onset of egg production is a major factor influencing hen productivity, as suitable body weight is crucial to laying performance in laying hens. To better understand the association between body weight and microbial community membership and structure in different sites of the digestive and reproductive tracts in chickens, we performed 16S rRNA sequencing surveys and focused on how the microbiota may interact to influence body weight. Our results demonstrated that the microbial community and structure of the digestive and reproductive tracts differed between low and high body weight groups. In particular, we found that the species Pseudomonas viridiflava was negatively associated with body weight in the 3 digestive tract sites, while Bacteroides salanitronis was negatively associated with body weight in the 3 reproductive tract sites; and further in-depth studies are needed to explore their function. These findings will help extend our understanding of the influence of the bird digestive and reproductive tract microbiotas on body weight trait and provide future directions regarding the control of body weight in the production of laying hens.
Key words: body weight, laying hen, microbiota, correlation analysis
INTRODUCTION
Animal phenotypic variation is non-negligibly affected not only by host genetics but also by numerous complex microorganisms and the environment (Nicholson et al., 2012; Martinez-Guryn et al., 2018). Many studies have shown that animals harbor a broad and complex microbiome that plays a vital role in nutrient absorption (Subramanian et al., 2014), disease resistance, environmental adaptation (Wang and Jia, 2016), health, physiology, and subsequent productive performance (Rosenberg and Zilber-Rosenberg, 2018). For instance, several studies have revealed that the composition of microbial communities at various anatomical locations in the animal body is influenced by diet, body weight (BW), early microbial exposure, and antibiotic use (Costello et al., 2009; Ottman et al., 2012; Knight et al., 2017).
As agricultural animals, chickens are an important type of domestic fowl that serve as both a major meat (broilers) and an egg source (laying hens), as well as an important animal model due to their small size and anatomical similarities to most birds (Burt, 2007). The digestive tract microbiota composition of chickens has been conclusively shown to be closely related to both host production and health (Yeoman et al., 2012; Singh et al., 2014; Kers et al., 2018; Khan and Moore, 2020; Yang et al., 2020). Numerous digestive tract microbiota-related studies have been conducted to investigate the relationship between the digestive tract microbiota and BW in broilers (Meng et al., 2014; Han et al., 2016; Lee et al., 2017; Johnson et al., 2018; Ji et al., 2019; Wen et al., 2019). Additionally, Bacteroides is the most abundant bacterial genus in lean chickens, but Clostridium is the most abundant bacterial genus in fat chickens (Ding et al., 2016; Hou et al., 2016; Xiang et al., 2021). However, most relevant published studies are based on broiler chickens. Identification and modulation of weight-related bacteria is one of the strategies to modulate BW, and it can potentially be a useful strategy to improve productive performance in the layers industry. Moreover, BW at the onset of egg production is a major factor influencing hen productivity, and suitable BW is crucial to laying performance in laying hens (Pérez-Bonilla et al., 2012a,b). Given that broilers and laying hens have major differences in their physiology, husbandry, feeding practices, and life span (Ocejo et al., 2019; Khan and Moore, 2020), it is necessary to study the digestive tract microbiota in laying hens with different BWs.
Recently, Wen et al. (2021a) used 16S rDNA analysis to characterize the microbial composition of 6 segments throughout the reproductive tract, including the cloaca, vagina, uterus, isthmus, magnum, and infundibulum. The authors found a relatively high abundance of vaginal Staphylococcus and Ralstonia that was significantly associated with darker eggshells. In addition, Lee et al. (2019) examined the microbial communities of the chicken oviduct and found a correlation between the maternal oviduct and chick gut microbiotas, suggesting vertical transmission of microbial communities present in the chicken oviduct during egg formation. The origin of the microbiota in the chicken reproductive tract has also been explored. Shterzer et al. (2020) found a large overlap in the composition of the gut (jejunum and cecum) and oviduct (infundibulum, magnum, and uterus) microbiotas that suggests transfer of material from the gut to the oviduct (Shterzer et al., 2020). These results demonstrate that the chicken oviductal microbiota has a close relationship with the digestive tract.
In poultry, feed moistened, reducing the coarseness of contents, nutrient digestion and absorption primarily occur in the crop, gizzard, and small intestine (including the duodenum, jejunum, and ileum), respectively (Rodrigues and Choct, 2018). These upper digestive tracts hold important digestive functions and play an important role in the performance and health of chickens (Rodrigues and Choct, 2018). The microbiome of the female reproductive tract benefits the chicken host due to the presence of antibacterial mechanisms, as previously suggested by Wen et al. (2021a). To further understand the effects of the digestive (crop, gizzard, and small intestine) and the reproductive (isthmus, uterus, and vagina) tracts microbiotas on BW, it is necessary to explore these mechanisms in laying hens.
In the current study, we performed 16S rRNA sequencing surveys and focused on the association between BW and microbial community membership and structure. We analyzed different sites of the digestive and reproductive tracts in chickens and evaluated how the microbiota interacts to influence BW. Using next-generation amplicon sequencing of the V4 region of the 16S rRNA gene, we sought to 1) characterize the microbiota of the crop, gizzard, small intestine, isthmus, uterus, and vagina within chickens with different BWs; 2) identify the contribution of key microbiota constituents at specified sites to BW; and 3) evaluate the microbiota at different sites positively/negatively associated with BW in chickens. Finally, we addressed the BW-related microorganisms and their correlations at the 6 sites.
MATERIALS AND METHODS
Ethics Statement
The complete procedure in this study was approved by the Committee on the Care and Use of Laboratory Animals of the State-Level Animal Experimental Teaching Demonstration Center of Sichuan Agricultural University.
Animals and Sample Collection
The Dongxiang Blue-shelled chicken is a Chinese indigenous breed that lays blue-shelled eggs with low egg productivity (Wang et al., 2007, 2009); this layer is characterized by black feathers and excellent egg quality that have a higher protein and lower cholesterol amount compared to commercial eggs (Wang et al., 2009). A total of 75 Dongxiang Blue-shelled layers from the Poultry Breeding Farm of Sichuan Agricultural University (Ya'an, China) were used in the present study. All fertilized chicken eggs hatched on the same day, and female chicks with similar body weights were selected on the final day of egg incubation. Subsequently, all hens were housed in neighboring pens in the same environment under a 16L:8D schedule with access to food and water ad libitum. No antibiotics were applied during the experimental period. In addition, the egg weight was measured at first egg and 300 d of age. The healthy chickens were sorted at the age of 300 d based on body weight and divided into 2 experimental groups: those with lower body weights (low body weight, LW; n = 37), and those with higher body weights (high body weight, HW; n = 38). Descriptive statistics of the body weight and egg production traits at first egg and 300 d of age in our experimental chickens are shown in Supplementary Table S1.
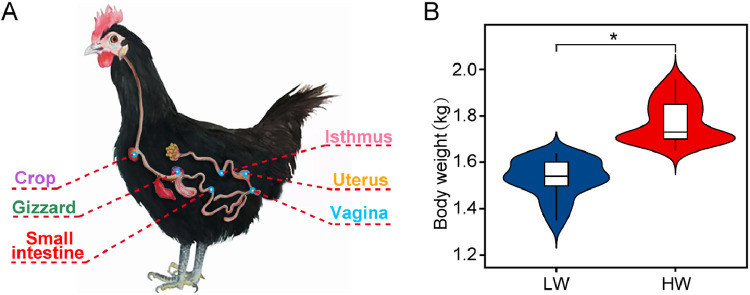
Subsequently, the 75 birds were humanely euthanized by cervical dislocation and all sample collection operations were aseptically performed on a clean bench. After the abdomen was opened, the viscera were removed, and the fresh tissues of the whole crop and gizzard were isolated. In addition, we also collected a 12-cm-long fixed midregion of the small intestine from each bird. For reproductive tract samples, the isthmus section was cut about 5-cm-long from the oviduct, and the whole uterus and vagina were also immediately sampled (Figure 1A). Next, the digestive segments were opened, the luminal mucosa was scraped from the region closest to the proventriculus to the region closest to the cloaca; the reproductive segments were opened, and the mucus was scraped with the back of a scalpel blade. All of the collected bird samples were snap frozen in liquid nitrogen and stored at −80°C prior to DNA extraction.
Figure 1.
Sampling sites and body weight in the chickens. (A) Spatial structure of the digestive and reproductive tracts of chickens and our six sampling sites. (B) Body weight comparison between low body weight (LW, n = 37) and high body weight (HW, n = 38) chickens. ∗P < 0.05.
DNA Extraction, 16S rRNA Gene Amplification and Sequencing
Microbial genomic DNA was extracted from the digestive and reproductive tract samples of each individual chicken using the commercially available TIANamp Stool DNA Kit (cat# DP328, TIANGEN, Beijing, China) in strict accordance with the manufacturer's instructions. The V4 region of the bacterial 16S rRNA gene was amplified by using the forward primer 515 F (5′-GTGCCAGCMGCCGCGGTAA-3′) and the reverse primer 806 R (5′-GGACTACHVGGGTWTCTAAT-3′) (Wen et al., 2021a). PCR amplifications were carried out by using Phusion High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA). The PCR products were purified using a GeneJET Gel Extraction Kit (Thermo Scientific, Schwerte, Germany). Library preparation was performed using an Ion Plus Fragment Library Kit 48 Rxns (Thermo Scientific, Schwerte, Germany) following the manufacturer's recommendations. Finally, sequencing was performed using an Ion S5 XL platform to generate 400 bp single-end reads at Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). The sequencing data have been deposited in the National Genomics Data Center (https://bigd.big.ac.cn/) (accession No. CRA002196).
Analysis of Sequencing Data
The raw reads were filtered to eliminate adapters and low-quality reads to obtain clean reads. For all samples, clean reads were clustered into operational taxonomic units (OTUs) via Uparse software (Uparse v7.0.1001) (http://drive5.com/uparse/) (Edgar, 2013) with a 97% sequence identity cutoff value. Unique sequences with ≥97% sequence similarity were assigned to the same OTUs. Subsequently, the Silva database (https://www.arb-silva.de/) (Quast et al., 2013; Yilmaz et al., 2014; Balvočiūtė and Huson, 2017) was used based on the Mothur algorithm to annotate taxonomic information for representative sequences. Subsequent analyses of alpha diversity indices, including ACE, Chao1, Good's coverage, observed species, PD whole tree, Shannon, and Simpson, were analyzed using the vegan package in R (version 2.15.3). Principal coordinate analysis (PCoA) and nonmetric multidimensional scaling (NMDS) were applied using R to estimate the similarity and discrepancies of bacterial communities among different sites based on the Bray-Curtis distance matrix. Linear discriminant analysis (LDA) effect size (LEfSe) (Segata et al., 2011) was performed to identify the bacterial taxa between LW and HW groups, with an LDA score >4 considered significant.
Statistical Analysis
The relationship between the abundance of phyla and BW was assessed by Pearson's correlation coefficient (r) and P values from simple linear regression. For the detection of microorganisms that were significantly associated with BW, we performed a statistical comparison between the highest 30% and lowest 30% BW-ranked chickens (2 distinct groups) using the Wilcoxon rank-sum test, and the association was considered significant if FDR <0.05. Subsequently, we calculated Pearson's and Spearman's correlation analyses between BW and the abundance of each microbiota at the genus and species levels using the psych package in R. We adjusted the P-value using the Benjamini-Hochberg (BH) approach (Benjamini and Hochberg, 1995). Correlations were considered significant if the adjusted P-value was lower than 0.05 (P < 0.05). Finally, the overlapping microorganisms obtained from the Wilcoxon rank-sum test, Spearman's and Pearson's correlations were considered to have a potential relationship with BW.
RESULTS
Sequencing Data
A total of 35,682,802 raw reads were yielded after 16S rRNA gene-based sequencing of all samples, and the data sets were then subjected to quality filtration procedures, which resulted in 33,646,292 clean reads for the subsequent analysis. The average number of sequences per sample was 74,770, and the number of sequences per sample ranged from 43,405 to 89,141 (Supplementary Table S2).
Microbiota Diversity According to BW and Sampling Site
We used PCoA and NMDS plots based on Bray–Curtis distances to compare the similarity between the compositions of the microbial communities in digestive and reproductive tracts. The digestive and reproductive tract samples (except small intestine) showed obvious separation, which means that the microbial communities in the digestive and reproductive tract samples have great differences (Supplementary Figure S1). To investigate the relationships between the digestive and reproductive tract microbiotas and BW traits in chickens, 75 chickens at the age of 300 d were divided into the LW and HW groups. The BW of the 2 groups at the age of 300 d showed a significant difference (P < 0.05; Figure 1B).
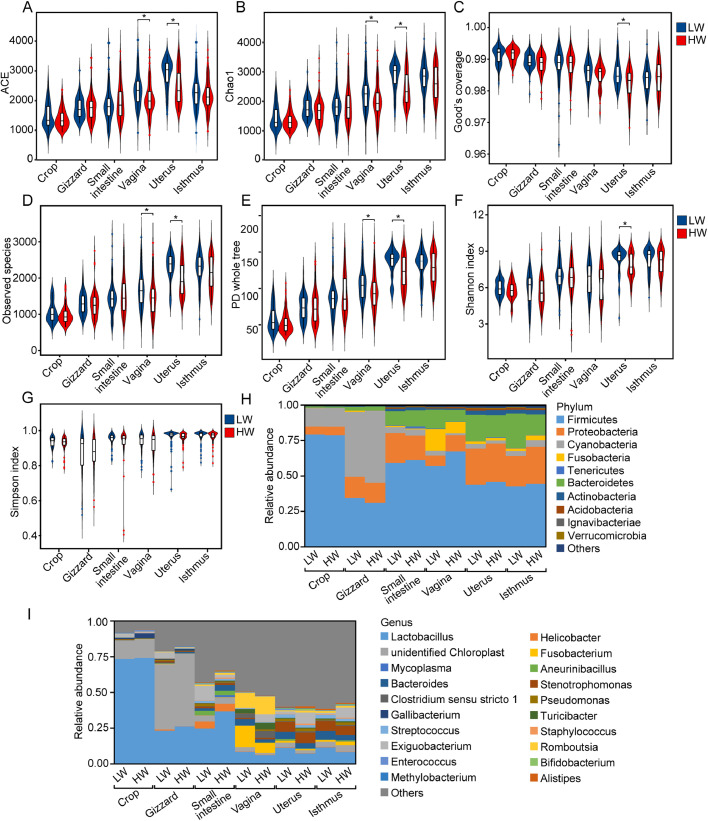
Next, various indices of alpha diversity were compared between the 2 groups, including the ACE, Chao1, Good's coverage, observed species, PD whole tree, Shannon, and Simpson indices, using OTUs at 97% identity. Interestingly, the comparison of ACE, Chao1, Good's coverage, observed species PD whole tree and Shannon indices of the 2 groups showed significant differences in the uterus (P < 0.05; Figures 2A–2F). Similarly, the comparison of ACE, Chao1, observed species, and PD whole tree of the 2 groups also showed significant differences in the vagina (P < 0.05; Figures 2A–2B and 2D–2E). Furthermore, the LW group had higher values for all indices than the HW group, although these indices were not significantly different between the 2 groups at the 6 sampling sites (P > 0.05; Figures 2A–2G). The results revealed that LW chickens have higher alpha diversity than HW chickens.
Figure 2.
Alpha diversity and microbial composition of digestive and reproductive tract microbiotas between LW and HW chickens. Alpha diversity comparison based on the ACE (A), Chao1 (B), good's coverage (C), observed species (D), PD whole tree (E), Shannon (F), and Simpson (G) indices, using the Wilcoxon rank-sum test to determine significant differences. ∗P < 0.05. Microbial composition of different BW groups at 6 sampling sites at the phylum (H), and genus (I) levels. Each bar represents the average relative abundance of each bacterial taxon within a group. Abbreviations: HW, high body weight; LW, low body weight.
Bacterial Community Composition and BW-Related Phyla
In the following work, we analyzed the bacterial community composition and structure of the digestive and reproductive tracts at different taxonomic levels (phylum and genus) using our sequencing data. At the phylum level, the 6 segments of the most common taxa were principally determined and belonged to Firmicutes, Proteobacteria, Cyanobacteria, Fusobacteria, and Bacteroidetes as the top 5 phyla. For instance, Firmicutes dominated the crop, small intestine, and vaginal microbiota, and the abundance of Firmicutes accounted for 78.93, 60.29, and 62.20% of the total abundance, respectively. However, Cyanobacteria represented the major proportion of the phyla in the gizzard microbiota, with a relative abundance of 48.19%. Interestingly, we found that 2 segments (uterus and isthmus) of the reproductive tract had similar dominant microorganism communities, which included Firmicutes (∼45%), Proteobacteria (∼24%), and Bacteroidetes (∼18%) (Figure 2H).
At the genus level, taxa unclassified below the family level were the most abundant in the three genital segments (vagina, uterus, and isthmus) (∼55%), followed by Lactobacillus, Fusobacterium, Bacteroides, and Stenotrophomonas (Figure 2I). We also found that 2 segments (uterus and isthmus) of the reproductive tract had similar dominant microorganism communities at the genus level, and such similarity was shown at the phylum level. However, Lactobacillus and unidentified Chloroplast were 2 dominant genera in the crop and gizzard, with a total combined abundance of 86.82 and 72.93% of the total abundance, respectively. Furthermore, Lactobacillus, unidentified Chloroplast, and Helicobacter represented the majority of the genera in the small intestine microbiota, with relative abundances of 30.70, 4.20, and 5.08%, respectively (Figure 2I). Relative taxon abundance plots at the genus level showed that the uterus and isthmus had similar microbial compositions.
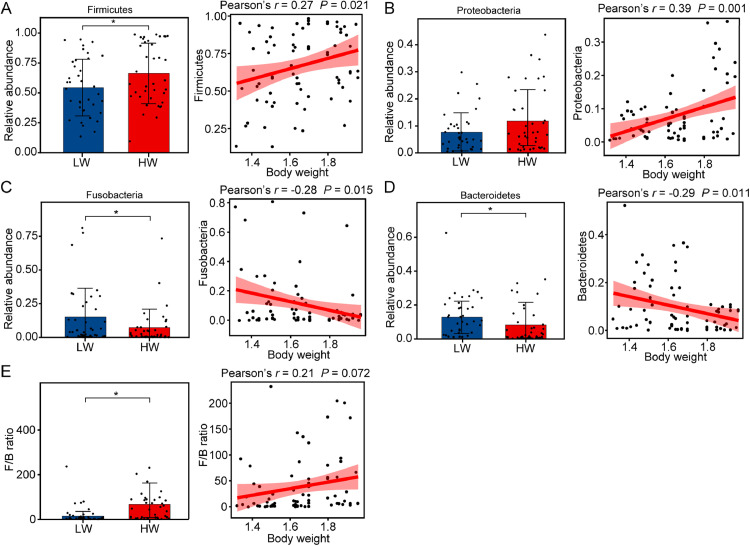
The dominant phylum Firmicutes was more abundant in the HW group than in the LW group (P < 0.05), and Firmicutes in the vagina was significantly positively correlated with BW (Pearson's r = 0.27, P = 0.021; Figure 3A). Meanwhile, Fusobacteria and Bacteroidetes in the vagina, which were more abundant in the LW group than in the HW group (P < 0.05), were significantly negatively associated with BW (Fusobacteria: Pearson's r = −0.28, P = 0.015; Bacteroidetes: Pearson's r = −0.29, P = 0.011; Figures 3C and 3D). The abundance of Proteobacteria in the vagina did not differ significantly between the HW and LW groups (P > 0.05) and was significantly positively correlated with BW (Pearson's r = 0.39, P = 0.001; Figure 3B). The Firmicutes/Bacteroidetes (F/B) ratio was significantly higher in the HW group than in the LW group (P < 0.05), but the F/B ratio was not significantly associated with BW (Pearson's r = 0.21, P = 0.072; Figure 3E).
Figure 3.
BW-related phyla in the vagina of chickens. Regression plots of BW against the relative abundance of the 4 dominant phyla. (A) Firmicutes, (B) Proteobacteria, (C) Fusobacteria, (D) Bacteroidetes, and (E) Firmicutes/Bacteroidetes (F/B) ratio; r: Pearson's correlation coefficient, *P < 0.05. Abbreviation: BW, body weight.
The BW-Specific Biomarkers in Six Sampling Sites
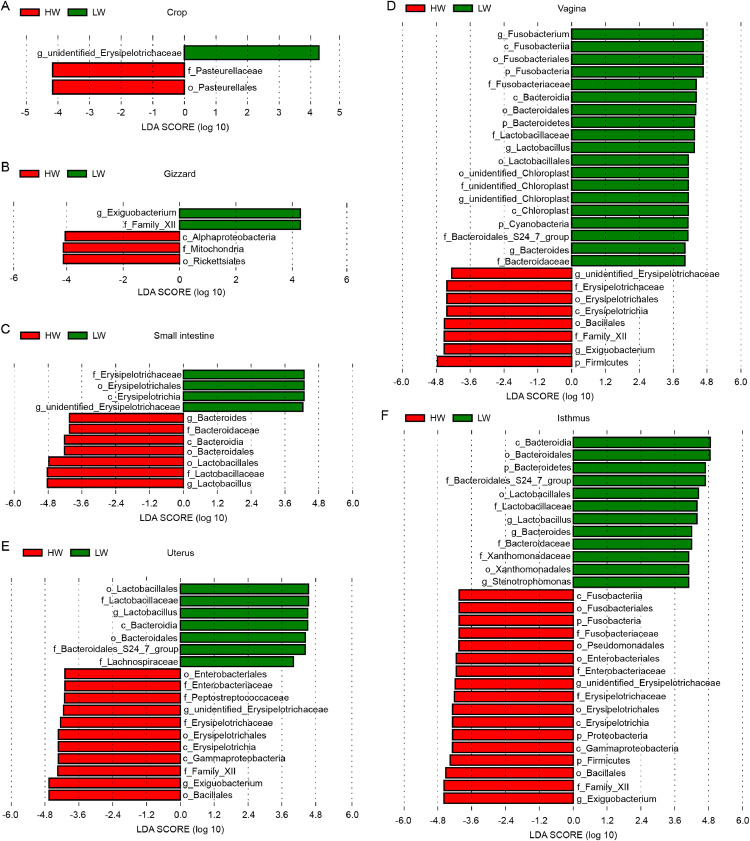
LEfSe was performed to identify specific taxa that varied in abundance between different BW groups and thus could be used as biomarkers. Three, 5, 11, 27, 18, and 29 bacterial taxa were significantly different between the LW and HW groups in the crop, gizzard, small intestine, vagina, uterus, and isthmus, respectively (LDA score >4; Figures 4A–4F). The 2 digestive segments (crop and gizzard) were associated with broad phylum-level changes in Firmicutes and Proteobacteria, and small intestine was associated with broad phylum-level changes in Firmicutes and Bacteroidetes. In contrast, the vagina, uterus, and isthmus were most prominently associated with broad phylum-level changes in Proteobacteria and Bacteroidetes, all of which were key phylotypes involved in the segregation of different BW groups in accordance with the LEfSe.
Figure 4.
BW-specific biomarkers in 6 sampling sites of chickens. LEfSe shows differentially abundant taxa as biomarkers determined using the Kruskal-Wallis test (P < 0.05), with an LDA score >4. Data presented were derived from (A) crop, (B) gizzard, (C) small intestine, (D) vagina, (E) uterus, and (F) isthmus samples. Abbreviations: BW, body weight; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis (LDA) effect size.
BW-Related Microorganisms and Their Correlations at the Six Sites
We further screened which microorganisms were indeed associated with BW. Only microorganisms that exhibited a significant correlation between BW and the relative abundance of the microbiota were considered causal. Subsequently, we found that 11 genera were significantly associated with BW, and 12 species were also associated with BW traits (P < 0.05). To visualize the relationship between BW and the microorganisms, Pearson's r values were calculated for BW with the relative abundance of the 11 genera and 12 species at the 6 sites. At the digestive tract sites, most microorganisms at the genus level were negatively correlated with BW (negative/positive: 11/3), whereas, in the reproductive tract sites, they were positively correlated with BW (negative/positive: 8/11) (Figure 5A). Notably, most species present in the 6 sites were negatively correlated with BW (negative/positive: 8/8 for the digestive tract sites; 12/7 for the reproductive tract sites) (Figure 5B).
Figure 5.
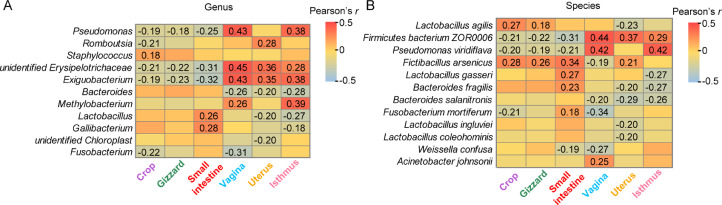
Microorganisms associated with BW. Pearson's r values between BW and BW-associated genera (11) (A) and species (12), (B) among the six sites. Red and blue tiles indicate positive and negative correlations, respectively. Significant r values are filled numerically (P < 0.05). Abbreviation: BW, body weight.
At the genus level, unidentified Erysipelotrichaceae and Exiguobacterium were negatively correlated with BW in the 3 digestive tract sites but positively correlated in the 3 reproductive tract sites (Figure 5A). Pseudomonas was negatively correlated with BW in the 3 digestive tract sites (Figure 5A and Supplementary Figure S2A). At the species level, Firmicutes bacterium ZOR0006 and Pseudomonas viridiflava in the 3 digestive tract sites were negatively correlated with BW (Figure 5B and Supplementary Figure S2B), but Firmicutes bacterium ZOR0006 in the 3 reproductive tract sites was positively correlated with BW. Fictibacillus arsenicus in the 3digestive tract sites had a significantly positive correlation with BW, while Bacteroides salanitronis in the 3 reproductive tract sites had a significantly negative correlation with BW (Figure 5B). Moreover, the LW chickens have a significantly higher abundance of Pseudomonas viridiflava and Bacteroides salanitronis compared to HW chickens in the 3 digestive and reproductive tract sites, respectively (P < 0.05; Supplementary Figure S3).
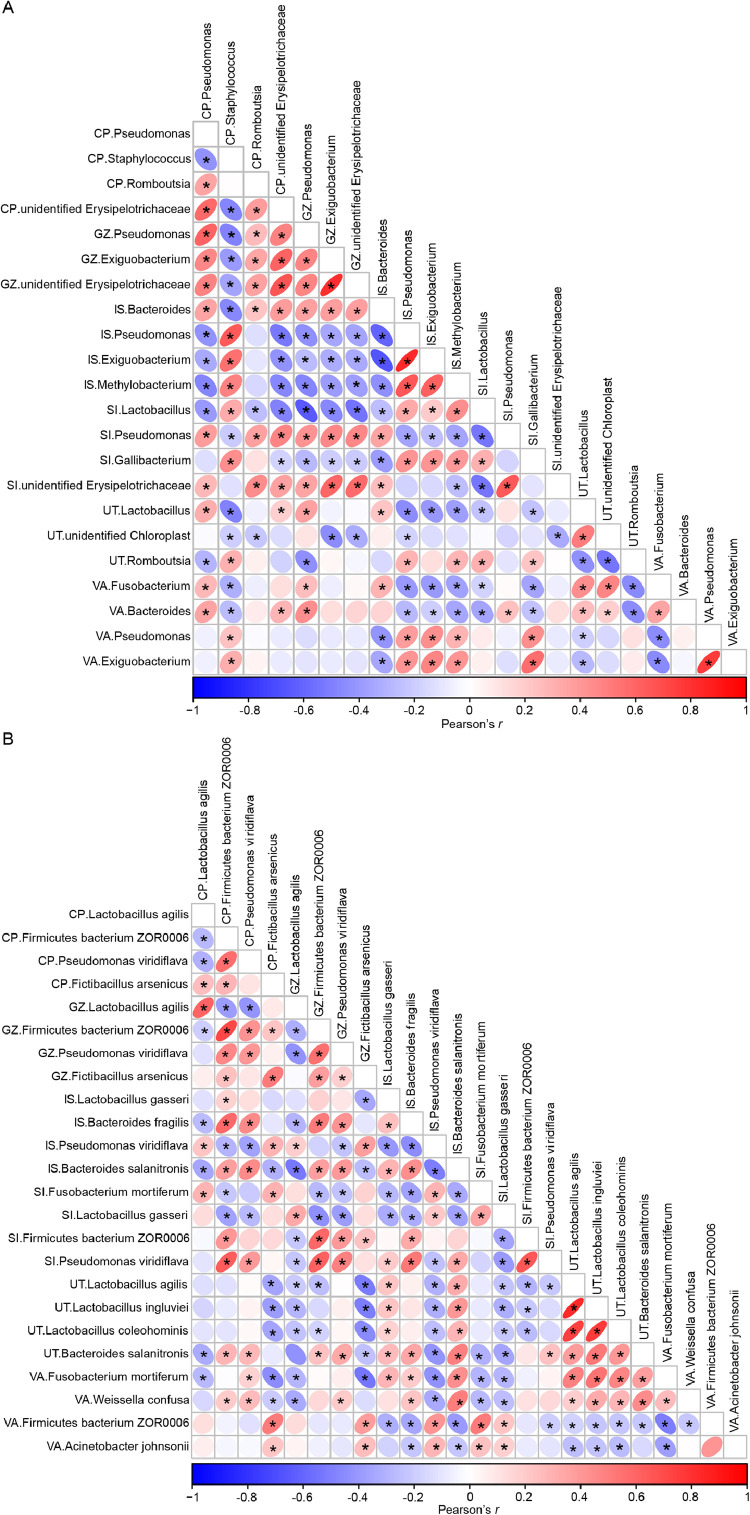
We subsequently focused on the relationships between these candidate microorganisms and each other at the genus and species levels. We found that the microorganisms in the uterus and isthmus were strongly correlated with each other, which implied a strong symbiotic relationship (Figure 6 and Supplementary Figure S4).
Figure 6.
BW-related microorganisms and their correlations. Pearson's r values of candidate microbial genera (A) and species (B) at the six sites. The intensity of the colors represents the degree of correlation, and the circle size represents the P value. Red and blue indicate positive and negative correlations, respectively; *P < 0.05. Abbreviations: BW, body weight; CP, crop; GZ, gizzard; IS, isthmus; SI, small intestine; UT, uterus; VA, vagina.
DISCUSSION
In this study, we observed that microbial communities were clearly separated in the digestive and reproductive tracts of laying hens. This result is consistent with previous reports that an obvious separation of community composition was seen among the crop, ileum, and cecum in chickens (Sekelja et al., 2012; Han et al., 2016). Growing evidence has confirmed that feed efficiency (Wen et al., 2021b), fat deposition (Wen et al., 2019; Xiang et al., 2021), and body weight (Han et al., 2016) are affected by the cecum microbial activity and composition in chickens, whereby the scope of our study is limited by the absence of cecum microbiota analysis. Importantly, there is clear evidence demonstrating that the microbial communities of the lower and upper reproductive tract are separated in laying hens (Wen et al., 2021a). In birds, given that the digestive and reproductive tracts share a common exit in the cloaca, the frequently exchanged microbiome likely resulted in similar microbiotas at the distal end of both tracts. Nonetheless, the small intestine microbiota was partially indistinguishable from that of the vagina. Previous studies have revealed that subjects with high BW have lower alpha diversity than subjects with low BW (Clarke et al., 2014; Han et al., 2016; Ramayo-Caldas et al., 2016; Panasevich et al., 2018; Oh et al., 2020). In our study, similar results were observed at the 6 sampling sites, although some indices were not significantly different between the LW and HW groups (P > 0.05). High alpha diversity is generally considered beneficial to animals (Reese and Dunn, 2018). However, a recent study has confirmed that limited diversity is more desirable and advantageous because not all microbes are beneficial (Foster et al., 2017; Reese and Dunn, 2018). Indeed, as described in previous studies, increasing diversity actually reduces stability in gut communities (Coyte et al., 2015). Hence, the decreased diversity observed in HW chickens might be due to an overabundance of a few dominant microorganisms and the presence of highly competitive microbial communities that may influence the richness and evenness of microbial community diversity (Bauer et al., 2018; Rodrigues et al., 2020).
Presently, it is well established that changes in intestinal microbial composition are an important causal factor in the development of obesity (Bäckhed et al., 2004; Turnbaugh et al., 2006; Bäckhed, 2009). In our study, we found that the relative abundance of Firmicutes gradually increased while that of Bacteroidetes decreased with increasing BW. This is in agreement with a previous study demonstrating that the 2 major bacterial phyla were associated with body weight in commercial chickens (Yadav et al., 2021). In addition to analyzing the relative abundance of the major phyla, we also found that vaginal samples from HW chickens had a significantly higher F/B ratio than samples from LW chickens. These observations are consistent with findings on mice (Turnbaugh et al., 2006), pigs (Mach et al., 2015; Han et al., 2017; Panasevich et al., 2018) and broiler chickens (Han et al., 2016) obesity. In particular, Western diet-induced obesity also caused an increase in the content of Fusobacteria and Proteobacteria. Previous studies found increased fecal Fusobacteria, which is a phylum that may be linked to proinflammatory stimuli to host epithelial cells and has been associated with colorectal carcinoma (Kostic et al., 2012; Andoh et al., 2016); however, the role of Fusobacteria in obesity is still largely unknown (Panasevich et al., 2018). In addition, Fusobacteria was more abundant in the LW group in our study, which is inconsistent with the findings. Overall, changes at the phylum taxonomic level from vaginal samples exhibited some consistency between chickens and what has been reported in the animals’ obesity literature. Further studies about the relationship between Fusobacteria and BW in various animals are needed to explain this result.
Lactobacillus and Exiguobacterium were found at 6 sites due to their broad adaptability or beneficial functions. Lactobacillus are thought to play key protective roles by lowering the environmental pH through lactic acid production (Boskey et al., 2001), so the high abundance of this genus in the digestive tracts corresponds to the low pH value of this organ system (Mabelebele et al., 2014). Lactobacillus or other lactic acid bacteria-dominant communities were less abundant in the uterus and isthmus, which is consistent with a recent study also performed in chickens (Lee et al., 2019). This observation may be related to the alkaline pH value of the chicken reproductive tract that helps maintaining sperm motility (Fiser and Macpherson, 1974; Mishra et al., 2018). Similar results also reported that the relative abundance of Lactobacillus accounts for less than 1% of the total microbiota community and that the pH of the vagina is nearly neutral in nonhuman primates (Yildirim et al., 2014), cows, pigs, and sheep (Miller et al., 2016). Exiguobacterium is a genus of bacilli and a member of the low-GC phylum Firmicutes; it has been found in areas covering a wide range of temperatures, pH values, salinity, and other conditions (White et al., 2018), and it also produces organic acids and hydrolytic enzymes (Kulshreshtha et al., 2012), even some having potential function in the degradation of toxic substances (Kasana and Pandey, 2018).
This study also enabled us to address some important biological questions regarding the chicken digestive and reproductive tract microbiotas in the LW and HW groups. BW-associated bacterial features in this study were identified by using LEfSe at the 6 sites. Interestingly, we observed phylum-level changes in Proteobacteria in the digestive and reproductive tracts, which were more abundant in the HW groups. In accordance with our findings, clear evidence from high-fat diet-fed mice (Zhang et al., 2010) demonstrated a strong association between increased proinflammatory gram-negative Proteobacteria and obesity, which may constitute a source of LPS causing systemic inflammation. Notably, Bacteroidetes bacterial taxa were detected as BW associated in the uterus and isthmus, and Bacteroides has an important role in food digestion and host immune function (Reeves et al., 1997). Therefore, these findings provide insights into the roles of the digestive and reproductive tract microbiota in chickens in improving protective host immunity to further increase BW.
Pseudomonas viridiflava, a member of the P. syringae, is phenotypically largely homogenous. The strains belonging to this species generally display a broad host range and can live either as pathogens or as saprophytes, playing an important role as animal and human pathogens (Bartoli et al., 2014; Samad et al., 2017). Thus, the prevalence of this species in the gut may associate to the observed decrease in the body weight of chickens. Our results demonstrated that Bacteroides salanitronis was negatively correlated with BW in reproductive tract sites. As an anaerobic bacterium in the gut, B. salanitronis is vital for N-glycan production (Nihira et al., 2013) and plays an important role in host obesity, immunodeficiency and diabetes (Zhao et al., 2019; Donaldson et al., 2020). Therefore, it is likely that B. salanitronis influences BW by regulating reproductive tract health in chickens. Our findings indicate that candidate microorganisms have a limited symbiotic relationship in digestive tract sites. Thus, in the production of laying hens, we could develop approaches to control BW by inhibiting the gut-associated Pseudomonas viridiflava abundance without altering the digestive and reproductive tract microbiota community.
In summary, our results demonstrated that the microbial community and structure of the digestive and reproductive tracts may affect BW traits through host-microbe interactions in laying hens. In particular, we found that the species Pseudomonas viridiflava was negatively associated with BW in the three digestive tract sites, while Bacteroides salanitronis was negatively associated with BW in the 3 reproductive tract sites; and further in-depth studies are needed to explore their function. These findings will help extend our understanding of the effects of the bird digestive and reproductive tract microbiotas on BW traits and provide future directions regarding the control of BW in the production of laying hens.
ACKNOWLEDGMENTS
This study was supported by the Sichuan Provincial Science and Technology Innovation Program (2020YFH0138).
DISCLOSURES
The authors declare no conflict of interest.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.psj.2021.101422.
Appendix. Supplementary materials
REFERENCES
- Andoh A., Nishida A., Takahashi K., Inatomi O., Imaeda H., Bamba S., Kito K., Sugimoto M., Kobayashi T. Comparison of the gut microbial community between obese and lean peoples using 16S gene sequencing in a Japanese population. J. Clin. Biochem. Nutr. 2016;59:65–70. doi: 10.3164/jcbn.15-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F. Changes in intestinal microflora in obesity: cause or consequence? J. Pediatr. Gastroenterol. Nutr. 2009;48:S56–S57. doi: 10.1097/MPG.0b013e3181a11851. [DOI] [PubMed] [Google Scholar]
- Bäckhed F., Ding H., Wang T., Hooper L.V., Koh G.Y., Nagy A., Semenkovich C.F., Gordon J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balvočiūtė M., Huson D.H. SILVA, RDP, greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genomics. 2017;18:114. doi: 10.1186/s12864-017-3501-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoli C., Berge O., Monteil C.L., Guilbaud C., Balestra G.M., Varvaro L., Jones C., Dangl J.L., Baltrus D.A., Sands D.C., Morris C.E. The Pseudomonas viridiflava phylogroups in the P. syringae species complex are characterized by genetic variability and phenotypic plasticity of pathogenicity-related traits. Environ. Microbiol. 2014;16:2301–2315. doi: 10.1111/1462-2920.12433. [DOI] [PubMed] [Google Scholar]
- Bauer M.A., Kainz K., Carmona-Gutierrez D., Madeo F. Microbial wars: competition in ecological niches and within the microbiome. Microb. Cell. 2018;5:215–219. doi: 10.15698/mic2018.05.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Stat. Methodol. 1995;57:289–300. [Google Scholar]
- Boskey E.R., Cone R.A., Whaley K.J., Moench T.R. Origins of vaginal acidity: high D/L lactate ratio is consistent with bacteria being the primary source. Hum. Reprod. 2001;16:1809–1813. doi: 10.1093/humrep/16.9.1809. [DOI] [PubMed] [Google Scholar]
- Burt D.W. Emergence of the chicken as a model organism: implications for agriculture and biology. Poult. Sci. 2007;86:1460–1471. doi: 10.1093/ps/86.7.1460. [DOI] [PubMed] [Google Scholar]
- Clarke S.F., Murphy E.F., O'Sullivan O., Lucey A.J., Humphreys M., Hogan A., Hayes P., O'Reilly M., Jeffery I.B., Wood-Martin R., Kerins D.M., Quigley E., Ross R.P., O'Toole P.W., Molloy M.G., Falvey E., Shanahan F., Cotter P.D. Exercise and associated dietary extremes impact on gut microbial diversity. Gut. 2014;63:1913–1920. doi: 10.1136/gutjnl-2013-306541. [DOI] [PubMed] [Google Scholar]
- Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyte K.Z., Schluter J., Foster K.R. The ecology of the microbiome: networks, competition, and stability. Science. 2015;350:663–666. doi: 10.1126/science.aad2602. [DOI] [PubMed] [Google Scholar]
- Ding J., Zhao L., Wang L., Zhao W., Zhai Z., Leng L., Wang Y., He C., Zhang Y., Zhang H. Divergent selection-induced obesity alters the composition and functional pathways of chicken gut microbiota. Genet. Sel. Evol. 2016;48:93. doi: 10.1186/s12711-016-0270-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson G.P., Chou W.C., Manson A.L., Rogov P., Abeel T., Bochicchio J., Ciulla D., Melnikov A., Ernst P.B., Chu H. Spatially distinct physiology of Bacteroides fragilis within the proximal colon of gnotobiotic mice. Nat. Microbiol. 2020;5:746–756. doi: 10.1038/s41564-020-0683-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Fiser P.S., Macpherson J.W. pH values in the oviduct of the hen during egg formation. Poult. Sci. 1974;53:827–829. doi: 10.3382/ps.0530827. [DOI] [PubMed] [Google Scholar]
- Foster K.R., Schluter J., Coyte K.Z., Rakoff-Nahoum S. The evolution of the host microbiome as an ecosystem on a leash. Nature. 2017;548:43–51. doi: 10.1038/nature23292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.G., Kim E.B., Lee J., Lee J.Y., Jin G., Park J., Huh C.S., Kwon I.K., Kil D.Y., Choi Y.J., Kong C. Relationship between the microbiota in different sections of the gastrointestinal tract, and the body weight of broiler chickens. Springerplus. 2016;5:911. doi: 10.1186/s40064-016-2604-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.G., Lee J.Y., Jin G.D., Park J., Choi Y.H., Chae B.J., Kim E.B., Choi Y.J. Evaluating the association between body weight and the intestinal microbiota of weaned piglets via 16S rRNA sequencing. Appl. Microbiol. Biotechnol. 2017;101:5903–5911. doi: 10.1007/s00253-017-8304-7. [DOI] [PubMed] [Google Scholar]
- Hou Q., Kwok L.Y., Zheng Y., Wang L., Guo Z., Zhang J., Huang W., Wang Y., Leng L., Li H., Zhang H. Differential fecal microbiota are retained in broiler chicken lines divergently selected for fatness traits. Sci. Rep. 2016;6:37376. doi: 10.1038/srep37376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J., Luo C.L., Zou X., Lv X.H., Xu Y.B., Shu D.M., Qu H. Association of host genetics with intestinal microbial relevant to body weight in a chicken F2 resource population. Poult. Sci. 2019;98:4084–4093. doi: 10.3382/ps/pez199. [DOI] [PubMed] [Google Scholar]
- Johnson T.J., Youmans B.P., Noll S., Cardona C., Evans N.P., Karnezos T.P., Ngunjiri J.M., Abundo M.C., Lee C.W. A consistent and predictable commercial broiler chicken bacterial microbiota in antibiotic-free production displays strong correlations with performance. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.00362-18. e00362-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasana R.C., Pandey C.B. Exiguobacterium: an overview of a versatile genus with potential in industry and agriculture. Crit. Rev. Biotechnol. 2018;38:141–156. doi: 10.1080/07388551.2017.1312273. [DOI] [PubMed] [Google Scholar]
- Kers J.G., Velkers F.C., Fischer E.A.J., Hermes G.D.A., Stegeman J.A., Smidt H. Host and environmental factors affecting the intestinal microbiota in chickens. Front. Microbiol. 2018;9:235. doi: 10.3389/fmicb.2018.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S., Moore R.J. The gut microbiota of laying hens and its manipulation with prebiotics and probiotics to enhance gut health and food safety. Appl. Environ. Microbiol. 2020;86:e00600–e00620. doi: 10.1128/AEM.00600-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight R., Callewaert C., Marotz C., Hyde E.R., Debelius J.W., McDonald D., Sogin M.L. The microbiome and human biology. Annu. Rev. Genomics Hum. Genet. 2017;18:65–86. doi: 10.1146/annurev-genom-083115-022438. [DOI] [PubMed] [Google Scholar]
- Kostic A.D., Gevers D., Pedamallu C.S., Michaud M., Duke F., Earl A.M., Ojesina A.I., Jung J., Bass A.J., Tabernero J., Baselga J., Liu C., Shivdasani R.A., Ogino S., Birren B.W., Huttenhower C., Garrett W.S., Meyerson M. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulshreshtha N.M., Kumar A., Bisht G., Pasha S., Kumar R. Usefulness of organic acid produced by Exiguobacterium sp. 12/1 on neutralization of alkaline wastewater. Sci. World J. 2012;2012 doi: 10.1100/2012/345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.C., Kil D.Y., Sul W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017;55:939–945. doi: 10.1007/s12275-017-7202-0. [DOI] [PubMed] [Google Scholar]
- Lee S., La T.M., Lee H.J., Choi I.S., Song C.S., Park S.Y., Lee J.B., Lee S.W. Characterization of microbial communities in the chicken oviduct and the origin of chicken embryo gut microbiota. Sci. Rep. 2019;9:6838. doi: 10.1038/s41598-019-43280-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mabelebele M., Alabi O.J., Ng`ambi J.W., Norris D., Ginindza M.M. Comparison of gastrointestinal tracts and pH values of digestive organs of Ross 308 broiler and indigenous venda chickens fed the same diet. Asian J. Anim. Vet. Adv. 2014;9:71–76. [Google Scholar]
- Mach N., Berri M., Estellé J., Levenez F., Lemonnier G., Denis C., Leplat J.J., Chevaleyre C., Billon Y., Doré J. Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 2015;7:554–569. doi: 10.1111/1758-2229.12285. [DOI] [PubMed] [Google Scholar]
- Martinez-Guryn K., Hubert N., Frazier K., Urlass S., Musch M.W., Ojeda P., Pierre J.F., Miyoshi J., Sontag T.J., Cham C.M., Reardon C.A., Leone V., Chang E.B. Small intestine microbiota regulate host digestive and absorptive adaptive responses to dietary lipids. Cell Host Microbe. 2018;23:458–469.e5. doi: 10.1016/j.chom.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H., Zhang Y., Zhao L., Zhao W., He C., Honaker C.F., Zhai Z., Sun Z., Siegel P.B. Body weight selection affects quantitative genetic correlated responses in gut microbiota. PLoS One. 2014;9:e89862. doi: 10.1371/journal.pone.0089862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E.A., Beasley D.E., Dunn R.R., Archie E.A. Lactobacilli dominance and vaginal pH: why is the human vaginal microbiome unique? Front. Microbiol. 2016;7:1936. doi: 10.3389/fmicb.2016.01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra A.K., Kumar A., Swain D.K., Yadav S., Nigam R. Insights into pH regulatory mechanisms in mediating spermatozoa functions. Vet. World. 2018;11:852–858. doi: 10.14202/vetworld.2018.852-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J.K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., Pettersson S. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–1267. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- Nihira T., Suzuki E., Kitaoka M., Nishimoto M., Ohtsubo K., Nakai H. Discovery of beta-1,4-D-mannosyl-N-acetyl-D-glucosamine phosphorylase involved in the metabolism of N-glycans. J. Biol. Chem. 2013;288:27366–27374. doi: 10.1074/jbc.M113.469080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocejo M., Oporto B., Hurtado A. 16S rRNA amplicon sequencing characterization of caecal microbiome composition of broilers and free-range slow-growing chickens throughout their productive lifespan. Sci. Rep. 2019;9:2506. doi: 10.1038/s41598-019-39323-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J.K., Chae J.P., Pajarillo E.A.B., Kim S.H., Kwak M.J., Eun J.S., Chee S.W., Whang K.Y., Kim S.H., Kang D.K. Association between the body weight of growing pigs and the functional capacity of their gut microbiota. Anim. Sci. J. 2020;91:e13418. doi: 10.1111/asj.13418. [DOI] [PubMed] [Google Scholar]
- Ottman N., Smidt H., De Vos W.M., Belzer C. The function of our microbiota: who is out there and what do they do? Front. Cell Infect. Microbiol. 2012;2:104. doi: 10.3389/fcimb.2012.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panasevich M.R., Wankhade U.D., Chintapalli S.V., Shankar K., Rector R.S. Cecal versus fecal microbiota in Ossabaw swine and implications for obesity. Physiol. Genomics. 2018;50:355–368. doi: 10.1152/physiolgenomics.00110.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Bonilla A., Jabbour C., Frikha M., Mirzaie S., Garcia J., Mateos G.G. Effect of crude protein and fat content of diet on productive performance and egg quality traits of brown egg-laying hens with different initial body weight. Poult. Sci. 2012;91:1400–1405. doi: 10.3382/ps.2011-01917. [DOI] [PubMed] [Google Scholar]
- Pérez-Bonilla A., Novoa S., García J., Mohiti-Asli M., Frikha M., Mateos G.G. Effects of energy concentration of the diet on productive performance and egg quality of brown egg-laying hens differing in initial body weight. Poult. Sci. 2012;91:3156–3166. doi: 10.3382/ps.2012-02526. [DOI] [PubMed] [Google Scholar]
- Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., Glöckner F.O. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y., Mach N., Lepage P., Levenez F., Denis C., Lemonnier G., Leplat J.J., Billon Y., Berri M., Doré J., Rogel-Gaillard C., Estellé J. Phylogenetic network analysis applied to pig gut microbiota identifies an ecosystem structure linked with growth traits. ISME. J. 2016;10:2973–2977. doi: 10.1038/ismej.2016.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese A.T., Dunn R.R. Drivers of microbiome biodiversity: a review of general rules, feces, and ignorance. mBio. 2018;9 doi: 10.1128/mBio.01294-18. e01294-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves A.R., Wang G.R., Salyers A.A. Characterization of four outer membrane proteins that play a role in utilization of starch by bacteroides thetaiotaomicron. J. Bacteriol. 1997;179:643–649. doi: 10.1128/jb.179.3.643-649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues D.R., Winson E., Wilson K.M., Briggs W.N., Duff A.F., Chasser K.M., Bielke L.R. Intestinal pioneer colonizers as drivers of ileal microbial composition and diversity of broiler chickens. Front. Microbiol. 2020;10:2858. doi: 10.3389/fmicb.2019.02858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues I., Choct M. The foregut and its manipulation via feeding practices in the chicken. Poult. Sci. 2018;97:3188–3206. doi: 10.3382/ps/pey191. [DOI] [PubMed] [Google Scholar]
- Rosenberg E., Zilber-Rosenberg I. The hologenome concept of evolution after 10 years. Microbiome. 2018;6:78. doi: 10.1186/s40168-018-0457-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad A., Antonielli L., Sessitsch A., Compant S., Trognitz F. Comparative genome analysis of the vineyard weed endophyte Pseudomonas viridiflava CDRTc14 showing selective herbicidal activity. Sci. Rep. 2017;7:17336. doi: 10.1038/s41598-017-16495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W.S., Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekelja M., Rud I., Knutsen S.H., Denstadli V., Westereng B., Næs T., Rudi K. Abrupt temporal fluctuations in the chicken fecal microbiota are explained by its gastrointestinal origin. Appl. Environ. Microbiol. 2012;78:2941–2948. doi: 10.1128/AEM.05391-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shterzer N., Rothschild N., Sbehat Y., Stern E., Nazarov A., Mills E. Large overlap between the intestinal and reproductive tract microbiomes of chickens. Front. Microbiol. 2020;11:1508. doi: 10.3389/fmicb.2020.01508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K.M., Shah T.M., Reddy B., Deshpande S., Rank D.N., Joshi C.G. Taxonomic and gene-centric metagenomics of the fecal microbiome of low and high feed conversion ratio (FCR) broilers. J. Appl. Genet. 2014;55:145–154. doi: 10.1007/s13353-013-0179-4. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Huq S., Yatsunenko T., Haque R., Mahfuz M., Alam M.A., Benezra A., DeStefano J., Meier M.F., Muegge B.D., Barratt M.J., VanArendonk L.G., Zhang Q., Province M.A., Petri W.A., Jr., Ahmed T., Gordon J.I. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature. 2014;510:417–421. doi: 10.1038/nature13421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Wang J., Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat. Rev. Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- Wang X.T., Deng X.M., Zhao C.J., Li J.Y., Xu G.Y., Lian L.S., Wu C.X. Study of the deposition process of eggshell pigments using an improved dissolution method. Poult. Sci. 2007;86:2236–2238. doi: 10.1093/ps/86.10.2236. [DOI] [PubMed] [Google Scholar]
- Wang X., Zheng J., Ning Z., Qu L., Xu G., Yang N. Laying performance and egg quality of blue-shelled layers as affected by different housing systems. Poult. Sci. 2009;88:1485–1492. doi: 10.3382/ps.2008-00417. [DOI] [PubMed] [Google Scholar]
- Wen C., Li Q., Lan F., Li X., Li G., Yan Y., Wu G., Yang N., Sun C. Microbiota continuum along the chicken oviduct and its association with host genetics and egg formation. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Yan W., Mai C., Duan Z., Zheng J., Sun C., Yang N. Joint contributions of the gut microbiota and host genetics to feed efficiency in chickens. Microbiome. 2021;9:126. doi: 10.1186/s40168-021-01040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen C., Yan W., Sun C., Ji C., Zhou Q., Zhang D., Zheng J., Yang N. The gut microbiota is largely independent of host genetics in regulating fat deposition in chickens. ISME. J. 2019;13:1422–1436. doi: 10.1038/s41396-019-0367-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R.A., 3rd, Soles S.A., Gavelis G., Gosselin E., Slater G.F., Lim D.S.S., Leander B., Suttle C.A. The complete genome and physiological analysis of the eurythermal firmicute exiguobacterium chiriqhucha strain RW2 isolated from a freshwater microbialite, widely adaptable to broad thermal, pH, and salinity ranges. Front. Microbiol. 2018;9:3189. doi: 10.3389/fmicb.2018.03189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang H., Gan J., Zeng D., Li J., Yu H., Zhao H., Yang Y., Tan S., Li G., Luo C. Specific microbial taxa and functional capacity contribute to chicken abdominal fat deposition. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.643025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav S., Caliboso K.D., Nanquil J.E., Zhang J., Kae H., Neupane K., Mishra B., Jha R. Cecal microbiome profile of Hawaiian feral chickens and pasture-raised broiler (commercial) chickens determined using 16S rRNA amplicon sequencing. Poult. Sci. 2021;100 doi: 10.1016/j.psj.2021.101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Liang S., Guo F., Ren Z., Yang X., Long F. Gut microbiota mediates the protective role of Lactobacillus plantarum in ameliorating deoxynivalenol-induced apoptosis and intestinal inflammation of broiler chickens. Poult. Sci. 2020;99:2395–2406. doi: 10.1016/j.psj.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeoman C.J., Chia N., Jeraldo P., Sipos M., Goldenfeld N.D., White B.A. The microbiome of the chicken gastrointestinal tract. Anim. Health Res. Rev. 2012;13:89–99. doi: 10.1017/S1466252312000138. [DOI] [PubMed] [Google Scholar]
- Yildirim S., Yeoman C.J., Janga S.C., Thomas S.M., Ho M., Leigh S.R., White B.A., Wilson B.A., Stumpf R.M., Consortium P.M. Primate vaginal microbiomes exhibit species specificity without universal Lactobacillus dominance. ISME. J. 2014;8:2431–2444. doi: 10.1038/ismej.2014.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P., Parfrey L.W., Yarza P., Gerken J., Pruesse E., Quast C., Schweer T., Peplies J., Ludwig W., Glöckner F.O. The SILVA and "All-species Living Tree Project (LTP)" taxonomic frameworks. Nucleic Acids Res. 2014;42:D643–D648. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Zhang M., Wang S., Han R., Cao Y., Hua W., Mao Y., Zhang X., Pang X., Wei C., Zhao G., Chen Y., Zhao L. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME. J. 2010;4:232–241. doi: 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- Zhao S., Lieberman T.D., Poyet M., Kauffman K.M., Gibbons S.M., Groussin M., Xavier R.J., Alm E.J. Adaptive evolution within gut microbiomes of healthy people. Cell Host Microbe. 2019;25:656–667. doi: 10.1016/j.chom.2019.03.007. e658. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.