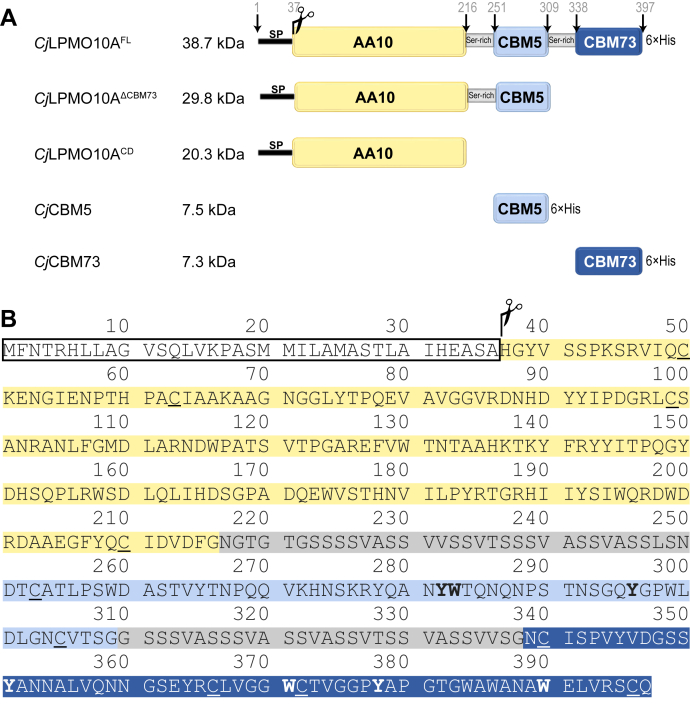
Figure 1.
Domain architecture and primary structure of CjLPMO10A.A, domain architecture and molecular weight of CjLPMO10A and the truncated variants used in this study. The numbers above the full-length enzyme show the transitions between the domains and the linkers. The signal peptide (residues 1–37) is cleaved off during secretion. The indicated molecular weights are based on the mature protein, that is, enzymes without signal peptides. B, primary structure of CjLPMO10AFL with color coding according to panel A. Aromatic residues located on the binding surfaces of the two CBMs, as determined in this study, are printed in bold face; cysteine residues involved in disulfide bonds are underlined. CBM, carbohydrate-binding module; CD, catalytic domain, Ser-rich linker; FL, full-length; His ×6, polyhistidine tag; SP, signal peptide.