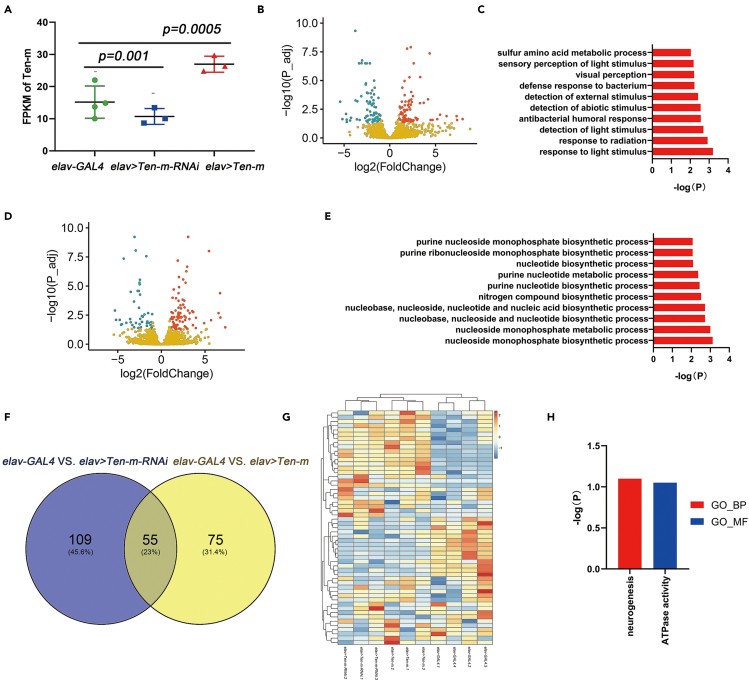
Figure 5.
RNA sequencing of brain tissues with aberrant expression of Ten-m in Drosophila
(A) FPKM of controls (elav-GAL4) and experimental flies (elav > Ten-m and elav > Ten-m-RNAi). Data are represented as mean ± SD. The Kruskal-Wallis test was used for comparison. Compared to the control groups, the experimental groups had increased or decreased the expression of Ten-m.
(B) Volcano plots of DEGs from elav > Ten-m-RNAi vs elav-GAL4 groups. Red denotes high expression while blue denotes low expression. In total, 164 significant DEGs were identified, including 83 upregulated genes and 81 downregulated genes.
(C) GO enrichment analysis of DEGs in the elav > Ten-m-RNAi group. The DEGs were enriched in stimulation-associated pathways, including light stimulus and detection of external stimulus.
(D) Volcano plots of DEGs from elav > Ten-m vs elav-GAL4 groups. Red denotes high expression while blue denotes low expression. In total, 130 significant DEGs were identified, including 85 upregulated genes and 45 downregulated genes.
(E) GO enrichment analysis of DEGs in the elav > Ten-m group. The DEGs were enriched in metabolism-associated pathways, such as the nucleoside monophosphate metabolic process.
(F) Venn diagram showing the intersection of significant DEGs observed in elav > Ten-m and elav > Ten-m-RNAi groups. There were 55 overlapped DEGs for both decreased and overexpressed Ten-m.
(G) Heatmap of overlapped significant DEGs. Hierarchical clustering was performed to highlight the DEGs, indicating a contrasting gene expression profile between experimental groups. Red denotes high expression while blue denotes low expression.
(H) GO enrichment analysis of overlapped significant DEGs in elav > Ten-m and elav > Ten-m-RNAi groups. The only BP term was neurogenesis, and the only MF term was ATPase activity.