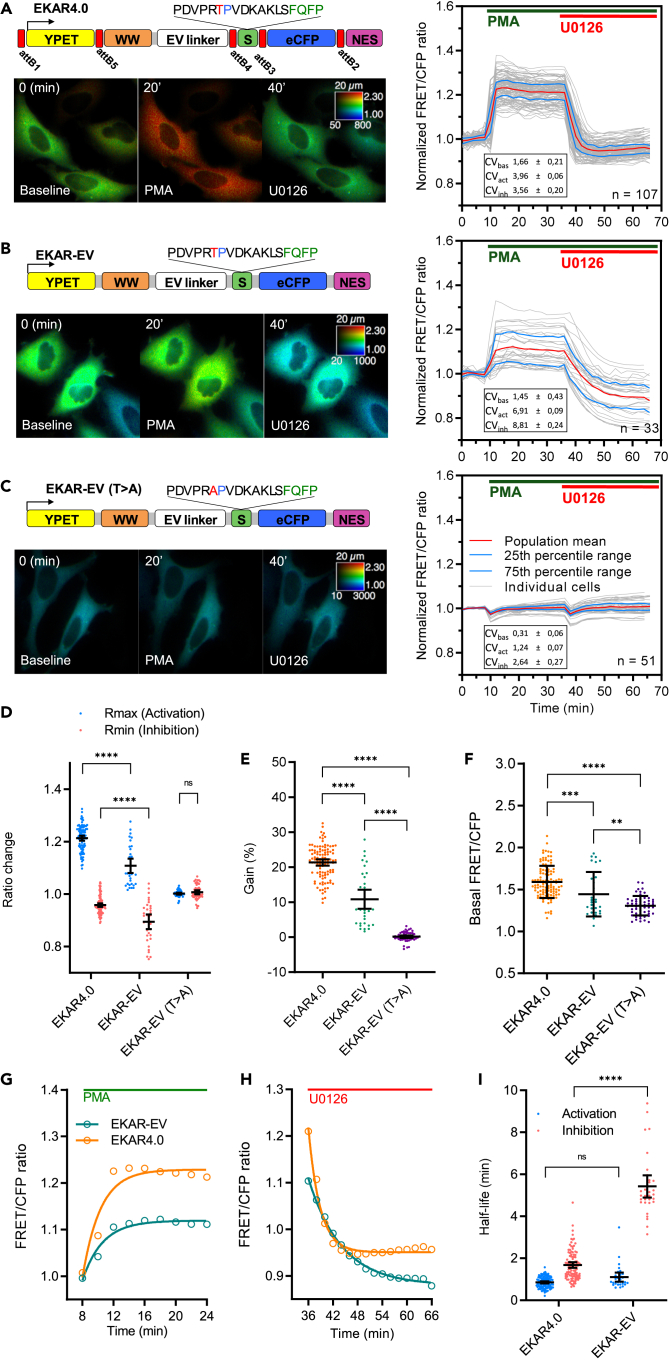
Figure 3.
Benchmarking EKAR4.0, our improved version of EKAR_EV biosensor, in living cells
(A–C) Biosensor reference experiments: HeLa cells expressing EKAR4.0 (A), EKAR-EV (B) or its mutant version, EKAR-EV (T > A) (C) were time lapse-imaged by FRET microscopy and treated with PMA (0.5 μM) at t = 10 min and then with U0126 (20 μM) at t = 34 min. Top of each panel: Schematic representation of ERK1/2 Kinase Activity Reporter (EKAR) construct used. The small red rectangles in (A) map the positions of obligated attB linkers between each FRET biosensor domain. For the substrate peptide sequence, red letters indicate the phosphorylation site, blue letters indicate amino-acid substitutions to increase the WW domain’s affinity, and green letters indicate ERK1/2 docking site. Bottom left of each panel: Pseudo-color FRET/CFP ratio images of ERK1/2 activity at the steady-state (t = 0 min), upon PMA addition (t = 20 min) and U0126 addition (t = 40 min) are shown. Right of each panel: Graph presenting the FRET/CFP ratio values of every single cell normalized to the averaged baseline FRET/CFP ratio value throughout reference experiments. All individual cells (grey curves), as well as the mean (red curve) and the 25th and 75th percentile range (blue curves) from at least 30 cells, are plotted as a function of time. CVbas, CVact, and CVinh indicate the coefficient of variation for cells at the baseline, cells in ERK1/2 activated state, and cells in the ERK1/2 inhibited state, respectively. Imaging parameters were kept identical for the different biosensors tested. Cells were imaged for a total duration of 70 min at the rate of 1 acquisition every 2 min.
(D–I) Characterization and comparison of different EKAR FRET biosensors responses were performed based on the following parameters: ratio changes (Rmin, Rmax) (D), gain (E), basal FRET/CFP value without any stimulation (F), and fitted curves to determine the speed of activation upon ERK1/2 activation (G, I) and the reversibility of the biosensor upon MEK1/2 inhibition (H, I). Fitted curves were plotted based on Tau ½, which is the time value representing half of the measured duration of the inhibition or activation phase. Tau ½ was calculated by applying a non-linear regression curve on ration values over the complete set of individual cells during the activation or inhibition phase. The nonlinear regression was calculated between t = 8 min and t = 24 min for the activation and between t = 36 min and t = 66 min for the inhibition and plotted as fitted curves. All calculations were performed using GraphPad PRISM 8. Data are mean ± SD from at least two independent experiments. Scale bar: 20 μm. The number of cells analyzed (n) is indicated on the lower right corner of the graph on the right panel (A–C). Statistical significance was determined using two-way ANOVA followed by Tukey’s post hoc test. Significance between samples is indicated as follows: ∗p< 0.05; ∗∗p< 0.01; ∗∗∗p< 0.001; ns, not significant.