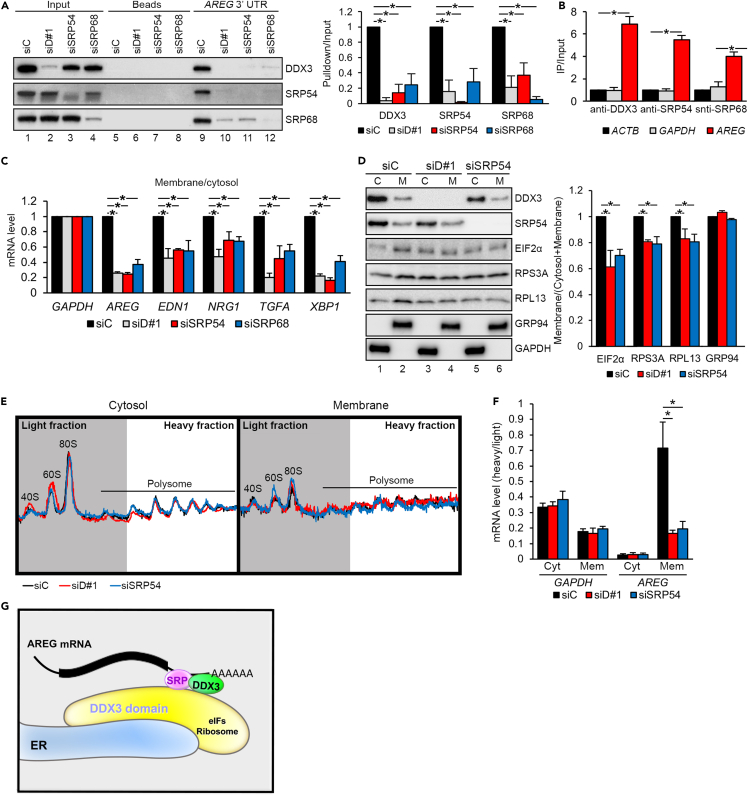
Figure 6.
DDX3 and SRP proteins are required for membrane association of target mRNAs
(A) Pull-down assay using the biotinylated AREG 3′ UTR RNA with lysates of siRNA-transfected SAS cells, followed by immunoblotting. Bar graph (mean ± SD; ∗p<0.01) shows relative levels of coprecipitation; siC was set to 1.
(B) RNA immunoprecipitation with SAS cell lysates using antibodies as indicated, followed by RT-qPCR analysis of the indicated mRNAs (mean ± SD; ∗p<0.01).
(C) RT-qPCR analysis of the indicated mRNAs in both the membrane and cytosol fractions of indicated siRNA-transfected SAS cells. Bar graph (mean ± SD; ∗p<0.01) shows the membrane-to-cytosol ratio of each mRNA in knockdown (siD#1, siSRP54, siSRP68) cells relative to control (siC).
(D) Immunoblotting of indicated proteins in the membrane and cytosol fractions from siRNA-transfected SAS cells. Bar graph shows the relative membrane/total (cytosol + membrane) ratio of the indicated proteins; siC was set to 1. Bars represent mean ± SD; ∗p < 0.01.
(E) Polysome profiles of the cytosol and membrane fractions of siRNA-transfected cells. Fractions from 40S to the first polysome (gray region) were collected as the light fractions; the following fractions (white region) were collected as the heavy fractions.
(F) RT-qPCR analysis of mRNAs from the heavy and light fractions of cytosol (Cyt) or membrane (Mem)-enriched polysomes. The heavy-to-light ratios of GAPDH and AREG in each sample were normalized to those of ACTB. ∗p < 0.01.
(G) DDX3 localizes proximally to the ER, where it associates with the SRP. DDX3/SRP cooperatively binds the 3′ UTR of target mRNAs and facilitates their translation by recruiting the translation machinery.