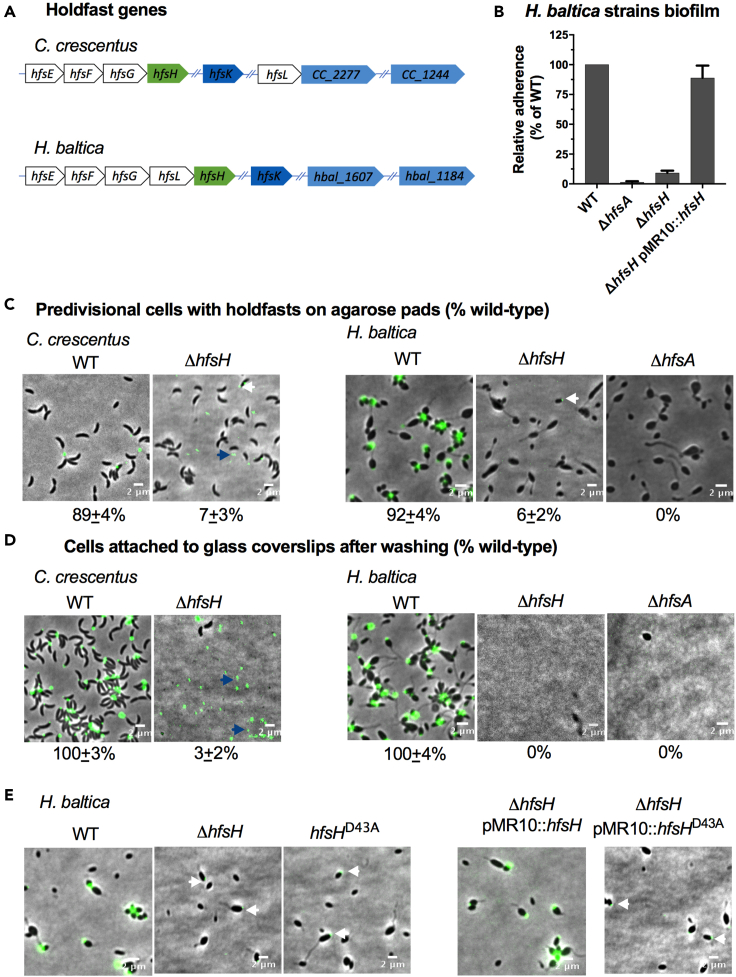
Figure 2.
The role of HfsH and HfsK in H. baltica holdfast biogenesis
(A) Genomic organization of holdfast synthesis (hfs) genes in C. crescentus and H. baltica. Genes were identified using reciprocal best hit analysis on C. crescentus and H. baltica genomes. In both C. crescentus and H. baltica genomes, hfsH is found in the hfs locus while hfsK and its paralogs are found outside the hfs locus. Color coding corresponds to homologs and paralogs. Hash marks indicate genes that are found in a different location in the genome.
(B) Quantification of biofilm formation by the crystal violet assay after incubation for 12 h, expressed as a mean percent of WT crystal violet staining. Holdfast null strain ΔhfsA was used as a negative control. Error is expressed as the standard error of the mean of three independent biological replicates with four technical replicates each.
(C) Representative images showing merged phase and fluorescence channels of the indicated C. crescentus and H. baltica strains on agarose pads. Holdfast is labeled with AF488-WGA (green). White arrows indicate holdfasts attached to the ΔhfsH cells, and blue arrows indicate holdfast shed into the medium. Scale bar, 2 μm. Exponential planktonic cultures were used to quantify the percentage of predivisional cells with holdfast. Data are expressed as the mean of three independent biological replicates with four technical replicates each. Error bars represent the standard error of the mean. A total of 3,000 cells were quantified per replicate using MicrobeJ.
(D) Representative images showing merged phase and fluorescence channels of C. crescentus and H. baltica strains bound to a glass coverslip. Exponential cultures were incubated on the glass slides for 1 h and washed to remove unbound cells, and holdfasts were labelled with AF488-WGA (green). Blue arrows indicate surface-bound holdfasts shed by hfsH mutants. Scale bar, 2 μm. The data showing quantification of cells bound to the glass coverslip are the mean of two biological replicates with five technical replicates each. Error is expressed as the standard error of the mean using MicrobeJ.
(E) Representative images showing merged phase and fluorescence channels of H. baltica strains with holdfast polysaccharides labeled with AF488-WGA (green) on agarose pads. Scale bar, 2 μm. A point mutation was introduced at a key substrate binding residue in H. baltica HfsH, resulting in an amino acid change from aspartic acid to alanine at position 43 (D43A). White arrows indicate faint AF488-WGA holdfast labeling on mutant cells