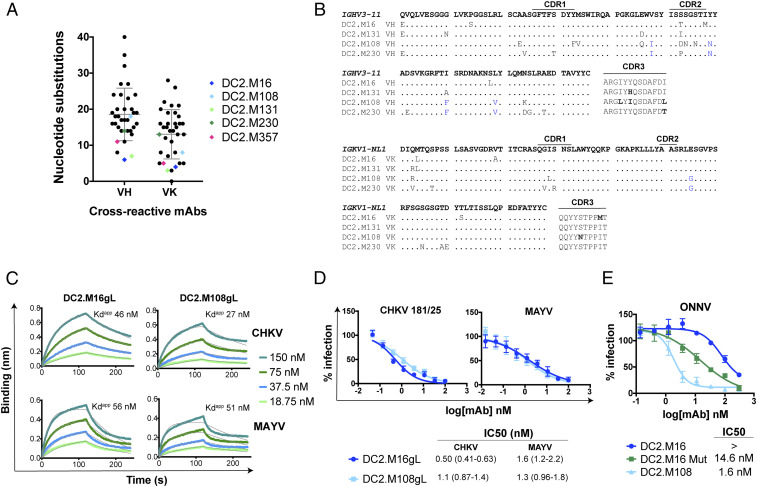
Fig. 6.
Germline sequence analysis and functional characteristics of inferred germline bNAbs. (A) Number of V-gene nucleotide substitutions for cross-reactive mAbs. bNAbs are highlighted as indicated. Mean and SD are shown in gray bars. (B) Alignment of mAbs DC2.M16, DC2.M131, DC2.M108, and DC2.M230 to the IGHV3-11/IGKV1-NL1 germline sequence. Mutations shared by DC2.M108 and DC2.M230 are highlighted in blue. (C) Binding of inferred germline mAbs DC2.M16gL and DC2.M108gL to CHIKV and MAYV p62-E1 proteins by BLI. (D) Neutralization of CHIKV and MAYV by inferred germline mAbs. Shown here are two independent experiments performed in triplicate wells (points represent mean ± SD). IC50 values were determined by fitting a nonlinear regression, and 95% confidence interval is shown. (E) Neutralization of ONNV by DC2.M16 after introduction of key mutations. Mutations shared by DC2.M108 and DC2.M230 were substituted into the DC2.M16 sequence (DC2.M16 Mut). Shown here are two independent experiments performed in triplicate wells (points represent mean ± SD). IC50 values were determined by fitting a nonlinear regression, and 95% CI is shown.