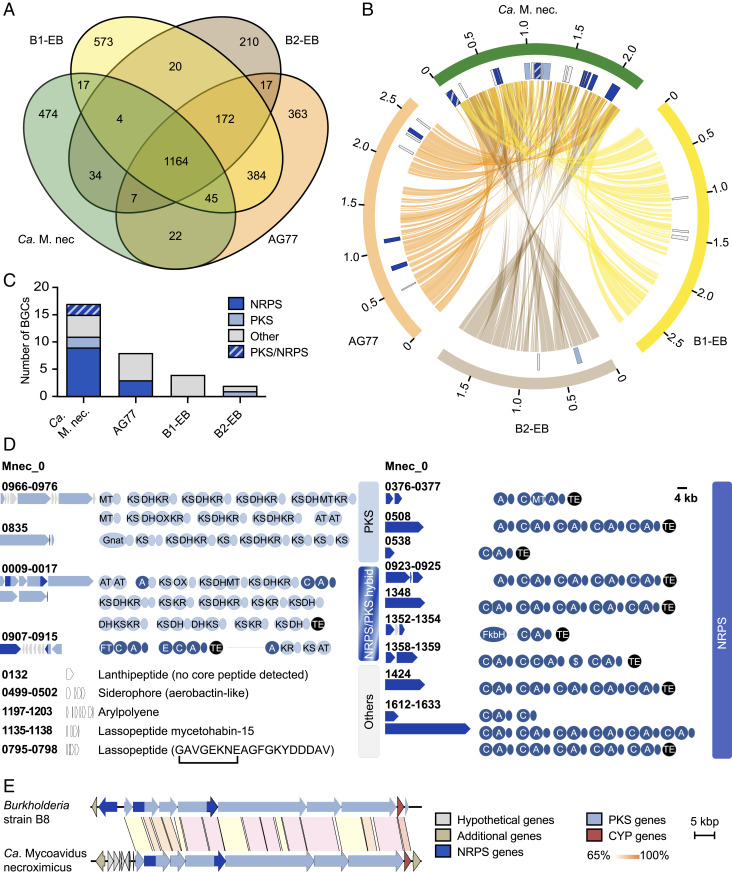
Fig. 2.
Comparative genomic analyses of Mycoavidus spp. (A) Number of orthologous proteins among the four Mycoavidus strains at 70% identity. (B) Circos plot of shared protein orthologs, and secondary metabolite loci (detected by antiSMASH v5) in Mycoavidus genomes. Outer blocks (orange, brown, yellow, green) represent genome sizes, while the inner blocks represent genomic positions of secondary metabolite loci. Lines linking the three genomes show position of genes whose proteins are orthologous at 70% identity. Depicted are the genome sequences of M. cysteinexigens strains AG77, B1-EB, B2-EB, and Ca. M. necroximicus (Ca. M. nec.). (C) Number of gene clusters putatively coding for natural products in Mycoavidus spp. detected by antiSMASH and by manual assignment. (D) BGCs and their encoded assembly lines identified from the endofungal Ca. M. necroximicus are displayed. A, adenylation; AT, acyltransferase; C, condensation; DH, dehydratase; E, epimerization; Gnat, GCN5-related N-acetyltransferase; KR, ketoreductase; KS, ketosynthase; MT, methyltransferase; OX, oxygenase; TE, thioesterase domains. Acyl carrier (light blue) and peptidyl carrier proteins (dark blue) are shown as circles without designators. (E) Homologous benzolactone BGCs in the genome of Burkholderia strain B8 and Ca. M. necroximicus.