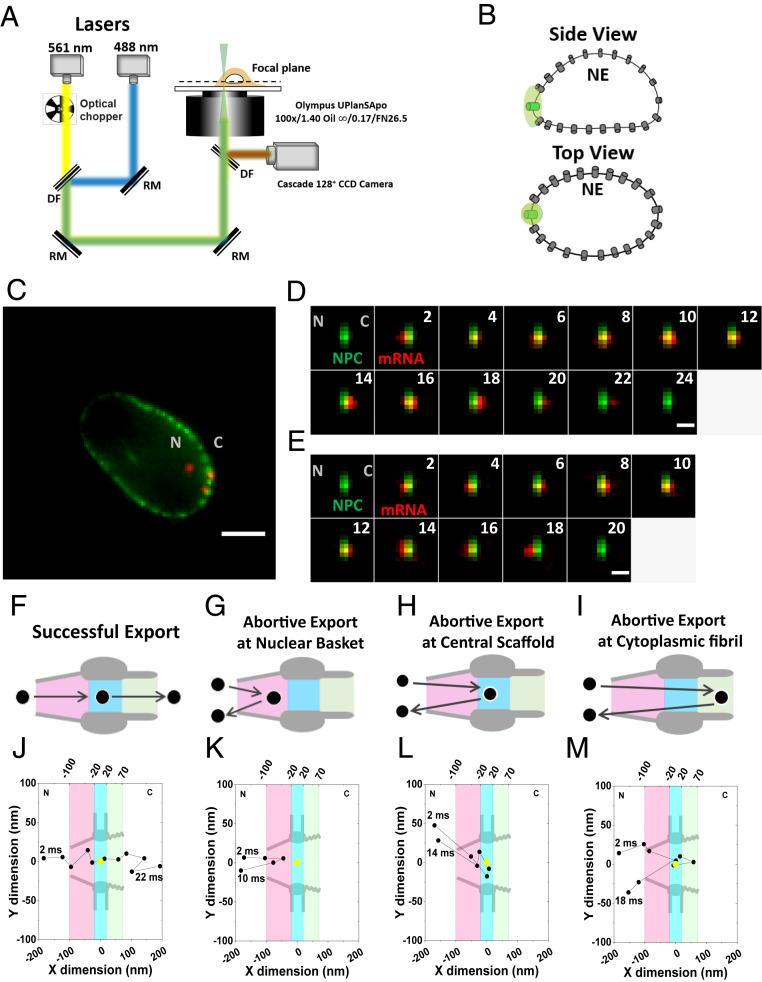
Fig. 2.
Nuclear export of mRNPs tracked by high-speed single-molecule SPEED microscopy. (A) Optical schematic of the high-speed single-molecule SPEED microscope. The 488-nm laser was utilized to excite NG-tagged NPCs at the equatorial plane of NE. The 561-nm laser was chopped by an optical chopper to achieve an on-off laser mode with a laser-on time of 60 ms and a laser-off time of 140 ms. The longer laser-off time gives enough time for fresh fluorescent mRNPs to diffuse from the nucleus into the NPC. (B) Individual NPC was illuminated by a point spread function of high-speed single-molecule SPEED microscopy at the equatorial plane of a DLD-1 cell nucleus in the focal plane. (C) An overlay of a wide-field epi-fluorescence image of the entire NE’s equator labeled with Nup153-NG (green curved line) and a narrow-field microscopy image of several mCherry-mRNPs particles (red spots) within a ∼3-μm area at the equator of the NE. (Scale bar: 5 μm.) C, cytoplasm; N, nucleus. (D) A typical successful single mRNP export event captured by high-speed single-molecule SPEED microscopy. A single mCherry-tagged mRNP (red spot) started from the nucleus, interacted with NPC (green spot), and arrived in the cytoplasm. Numbers denote time in milliseconds. (Scale bar: 1 μm.) C, cytoplasm; N, nucleus. (E) A typical abortive single mRNP export event. A single mCherry-tagged mRNP (red spot) started from nucleus, interacted with NPC (green spot), and returned to the nucleus. Numbers denote time in milliseconds. (Scale bar: 1 μm.) C, cytoplasm; N, nucleus. (F) The model of successful mRNPs export. The mRNP (black circle) sequentially diffuses through nuclear basket (pink region), central scaffold (light blue region), and cytoplasmic fibril (light green region) of the NPC into cytoplasm. (G–I) The models for three types of mRNP’s abortive export. (J) A typical successful export trajectory of mRNA. Single-particle tracks (black dots) were acquired by 2D Gaussian fitting to point spread functions of mRNP–mCherry particles in a series of images for the successful event in D. The centriole of NPC (yellow dot) was acquired by 2D Gaussian fitting to point spread function of single NPC-NG in D. Based on NPC scaffold structure obtained from EM study, nuclear basket (pink region), central scaffold (light blue region), and cytoplasmic fibril (light green region) along the axial dimension are arbitrarily defined as 80 nm, 40 nm, and 50 nm in length, respectively. C, cytoplasm; N, nucleus. (K–M) The examples of three types of mRNA abortive export trajectories. C, cytoplasm; N, nucleus.