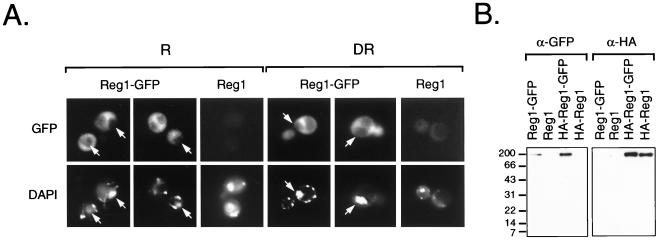
FIG. 4.
Subcellular localization of Reg1-GFP. (A) Fluorescence micrographs of KDY82 cells expressing Reg1-GFP. Cells expressing Reg1-GFP as the sole source of functional Reg1 were created by transforming strain KDY82 with plasmid pKD109. KDY82 expressing HA-Reg1 from plasmid pKD89 served as the negative control for yeast cell autofluorescence. Repressed cells (R) were grown in medium containing 5% glucose. A portion of these cells was derepressed (DR) by shifting them to medium containing 0.05% glucose. The derepressed cells shown in these micrographs were prepared 4 h after the shift. By this time, ADH2-lacZ expression was beginning to derepress and SUC2 expression had fully derepressed (data not shown). Cells examined at 2, 4, 8, and 12 h after the shift showed the same distribution of GFP fluorescence. Arrows in the micrographs mark the position of nuclei based on the location of DAPI fluorescence. (B) Western blot analysis of Reg1-GFP. Cell extracts were prepared from repressed KDY82 transformants expressing Reg1-GFP from plasmid pKD109, Reg1 from plasmid pKD63, HA-Reg1-GFP from plasmid pKD110, and HA-Reg1 from plasmid pKD89. Proteins from 100 μg of each cell extract were separated on an SDS–12% acrylamide gel and transferred to nitrocellulose. GFP-tagged proteins were detected with anti-GFP polyclonal antibodies (α-GFP), and HA-tagged proteins were detected with anti-HA monoclonal antibodies (α-HA).