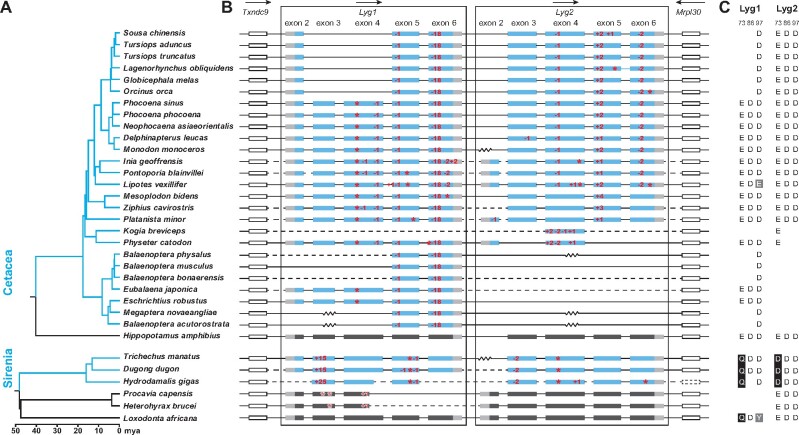
Fig. 1.
Loss of Lyg genes in aquatic mammals based on published sequences. (A) Species trees for Cetacea and Sirenia (blue clades), and their close relatives (black clades), with divergence time shown (Upham et al. 2019). (B) Coding regions of the Lyg1 and Lyg2 genes from each species is shown, which are located between the Txndc9 and Mrpl30 genes. For aquatic mammals, exons for the coding regions of Lyg1 and Lyg2 are shown in blue, with untranslated regions in gray. The two flanking genes are represented by hollow rectangles, with the dotted one showing a missing gene. Only exons are drawn to scale, with introns indicated by horizontal lines. The zig-zag line represents a gap in the genomic sequence within a scaffold, whereas the dashed line indicates a gap as the sequences are from different scaffolds. Arrows above the genes indicate the direction of gene transcription. Frameshift indels and premature stop codons are indicated in red. Indels with lengths that are multiples of 3, but not more than nine bases long (three amino acids), are not marked in the exons. (C) Numbering of the key catalytic amino acid residues 73, 86, and 97 are based on the goose Lyg positions, with the substitutions shaded in either black (for site 73) or gray (for both 86 and 97).